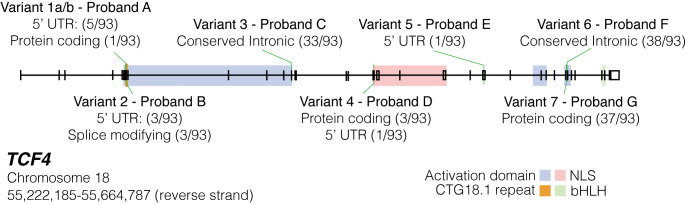
Fig 5. Schematic map of rare and potentially deleterious variants identified in individuals with molecularly unsolved FECD within TCF4, without CTG18.1 expansions.
All 8 rare (minor allele frequency (MAF)<0.005) and potentially pathogenic (CADD>15) variants in 7 Exp- Probands are mapped along TCF4 genomic region and transcriptomic locations contextualised. Proband A variants are found in cis within the proband. Conserved intronic transcripts refer to variants located in intronic regions identified as conserved via Genomic Evolutionary Rate Profiling (GERP) conservation scores on Ensembl. Splice modifying refers to transcripts where the variant identified would induce alternative splicing. Transcript numbers where the variant occurred within introns were not included. UTR: untranslated region. NLS: nuclear localization signal. bHLH: basic helix loop helix.