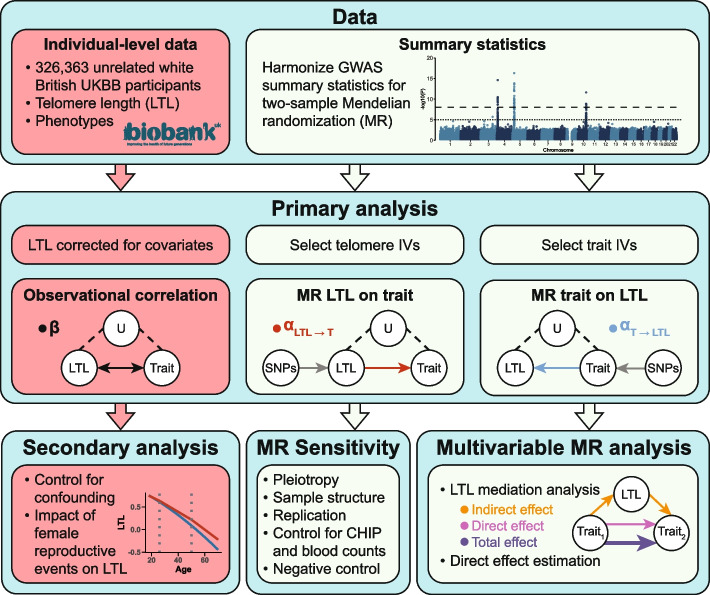
Fig. 1.
Schematic representation of the study’s workflow. Red and light green boxes denote steps using individual-level phenotypic data from the UK Biobank and genome-wide association studies (GWAS) summary statistics, respectively. Top: Data extraction process. Middle: Analyses focused on leukocyte telomere length (LTL) trait relationships including observational correlation (; black), Mendelian randomization (MR) to assess the impact of LTL on traits (; red), and MR to assess the impact of traits on LTL (; blue). LTL covariates comprise age, age2, genotyping array, sex, and the interaction of the latter with the priors. U = unmeasured confounding factors; IVs = instrumental variables, i.e., genetic variants with genome-wide significant association to the considered trait. SNPs = single nucleotide polymorphisms. Bottom: Follow-up analyses include exploring the association of female reproductive events with LTL, sensitivity analyses (i.e., implementation of seven complementary MR methods, replication using independent LTL summary statistics [18], controlling for confounding by blood-related traits, and evaluating the MR effect of LTL on sex as a negative control), and perform mediation analysis through multivariable MR. Note that for mediation analyses, both exposure and mediator are instrumented. CHIP = clonal hematopoiesis of indeterminate potential