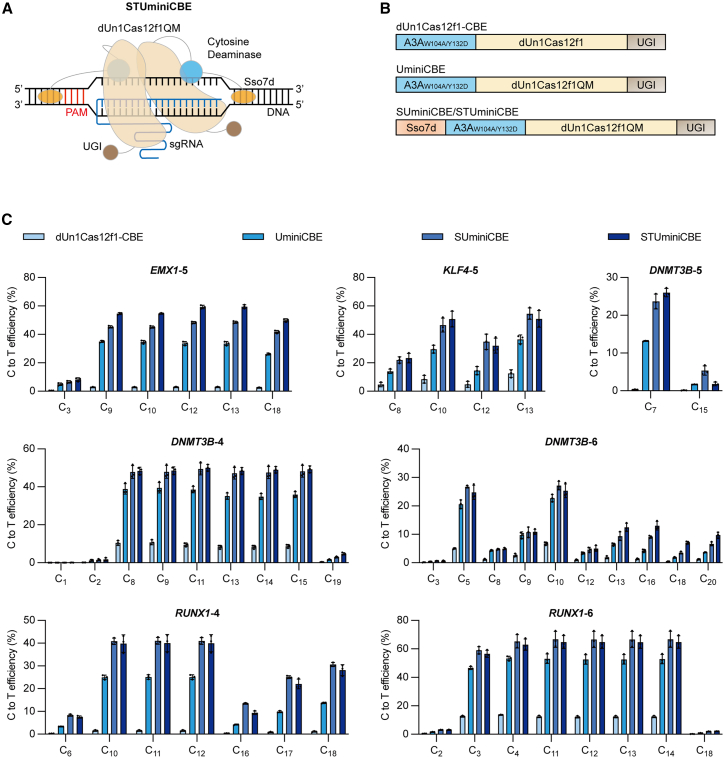
Figure 2.
Development of dUn1Cas12f-mediated cytosine base editors
(A) Schematic of nuclease-dead Un1Cas12f1-derived miniature cytosine base editor (STUminiCBE). STUminiCBE consists of dUn1Cas12f1QM (light orange) fused with cytosine deaminase (light blue) and Sso7d (orange) at the N terminus and UGI (brown) at the C terminus, along with truncated sgRNA (blue) for DNA targeting and cytosine base editing. (B) Diagram of dUn1Cas12f1-CBE, UminiCBE, SUminiCBE, and STUminiCBE. All CBEs consist of the cytosine deaminase A3AW104A/Y132D (light blue) at the N terminus, dUn1Cas12f1 or dUn1Cas12f1QM (light orange) in the middle, and UGI (brown) at the C terminus. SUminiCBE or STUminiCBE contains Sso7d (orange) at the N terminus of A3AW104A/Y132D-dUn1Cas12f1QM-UGI. Original sgRNA and truncated sgRNA-D1/D2/D3 are not shown in the diagram. (C) Comparison of C-to-T editing efficiencies mediated by dUn1Cas12f1-CBE, UminiCBE, SUminiCBE, and STUminiCBE at EMX1-5, KLF4-5, DNMT3B-4, DNMT3B-5, DNMT3B-6, RUNX1-4, or RUNX1-6 site. All values and error bars represent means ± SD, n = 3 independent biological replicates.