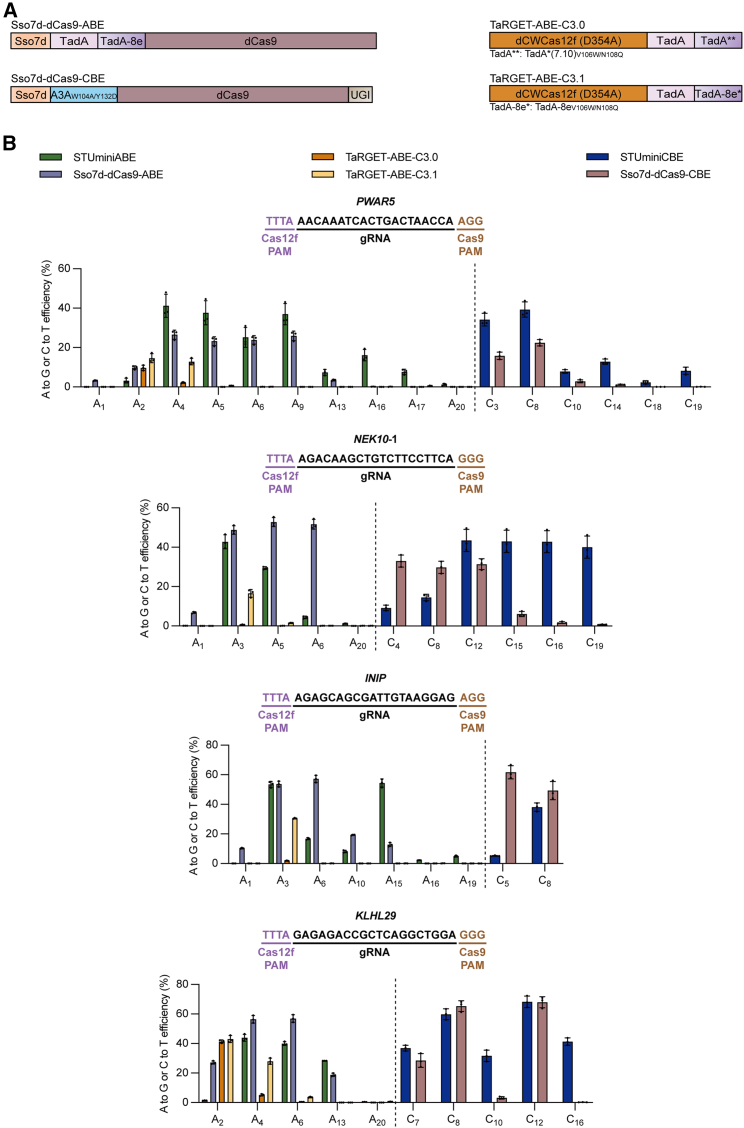
Figure 3.
Comparison of the base-editing efficiencies induced by STUminiBEs, dCas9-derived BEs, or TaRGET-ABEs
(A) Diagram of Sso7d-dCas9-ABE, Sso7d-dCas9-CBE, TaRGET-ABE-C3.0, and TaRGET-ABE-C3.1. Sso7d-dCas9-ABE consists of Sso7d (orange) at the N terminus, adenine deaminases (TadA is in pink, TadA-8e is in purple) in the middle, and dead SpCas9 (purple brown) at the C terminus. Sso7d-dCas9-CBE contains Sso7d and the cytosine deaminase A3AW104A/Y132D (light blue) at the N terminus, dCas9 in the middle, and UGI (brown) at the C terminus. TaRGET-ABE-C3.0 and TaRGET-ABE-C3.1 both consist of dCWCas12f (D354A) (dark orange) at the N terminus and adenine deaminases at the C terminus. TaRGET-ABE-C3.0 contains TadA∗∗ that denotes the engineered form with V106W and N108Q mutations in Tad∗(7.10), while TaRGET-ABE-C3.1 contains TadA-8e∗ that denotes the engineered form with V106W and N108Q mutations in TadA-8e. sgRNA is not shown in the diagram. (B) Comparison of A-to-G editing efficiencies mediated by STUminiABE, Sso7d-dCas9-ABE, TaRGET-ABE-C3.0, and TaRGET-ABE-C3.1 and C-to-T editing efficiencies mediated by STUminiCBE and Sso7d-dCas9-CBE at PWAR5, NEK10-1, INIP, or KLHL29 site. dUn1Cas12f1 or dCWCas12f PAM sequences are in light purple, and dCas9 PAM sequences are in brown. All values and error bars represent means ± SD, n = 3 independent biological replicates.