Highlights
-
•
The NKp30 receptor's expression on activated NK cells is more pronounced in DF patients than in DHF patients, suggesting a potential role of this receptor in the milder disease manifestation observed in pediatric dengue patients.
-
•
Natural killer cells are activated upon engagement with dengue infected dendritic cells and the outcome of cellular activation depends on the type of receptor expressing on NK cells and the intracellular signaling pathway.
-
•
The NK cells response to the dengue virus operates through distinct mechanisms. Degranulation is initiated by direct interaction of NK cells and dengue infected cells and signaling from type I IFNs. The production of IFN-γ is first initiated by direct cell-cell contact and type i IFNs followed by several cytokines including type I IFNs, IL-12, IL-15, and IL-18.
Keywords: Dengue virus, NK cells, DHF, Innate immunity, NKp30
Abstract
Natural killer cells (NK cells) are the front line of immune cells to combat pathogens and able to influence the subsequent adaptive immune responses. One of the factors contributing to pathogenesis in dengue hemorrhagic fever (DHF) disease is aberrant immune activation during early phase of infection. This study explored the profile of NK cells in dengue infected pediatric patients with different degrees of disease severity. DHF patients contained higher frequency of activated NK cells but lower ratio of CD56dim:CD56bright NK subsets. Activated NK cells exhibited alterations in several NK receptors. Interestingly, the frequencies of NKp30 expressing activated NK cells were more pronounced in dengue fever (DF) than in DHF pediatric patients. In vitro functional analysis indicated that degranulation of NK cells in responding to dengue infected dendritic cells (DCs) required cell-cell contact and type I IFNs. Meanwhile, Interferon gamma (IFN-γ) production initially required cell-cell contact and type I IFNs followed by Interleukin-12 (IL-12), Interleukin-15 (IL-15) and Interleukin-18 (IL-18) resulting in the amplification of IFN-γ producing NK cells over time. This study highlighted the complexity and the factors influencing NK cells responses to dengue virus. Degree of activation, phenotypes of activated cells and the crosstalk between NK cells and other immune cells, could modulate the outcome of NK cells function in the dengue disease.
1. Introduction
The increasing rate of dengue virus (DENV) transmission in tropical and subtropical areas around the world has raised global public health concern. There has been estimated that up to 390 million people worldwide are infected with dengue virus annually and among these, 96 million people develop dengue disease (Bhatt et al., 2013). Dengue virus is transmitted to humans through the bite of infected mosquitoes resulting in a wide spectrum of dengue diseases, ranging from asymptomatic, mild (dengue fever, DF) to severe (dengue hemorrhagic fever, DHF) diseases. The mortality rate of DHF patients can be as high as 20 % without the appropriate care and there is no specific drug available (World Health Organization, 1997). Dengue virus infection can trigger multiple components of immunity. Studies in dengue infected patients demonstrated the high viral load together with high levels of immune activation during febrile illness including massive activation of dengue specific T cells, elevated levels of serum cytokines or chemokines such as IFN-α, TNF-α, MIP-1β and IP-10 (Bozza et al., 2008; Hober et al., 1993; Kurane et al., 1993; Mongkolsapaya et al., 2003; Vaughn et al., 2000). Dengue virus-specific antibodies are able to enhance disease severity through antibody dependent enhancement mechanisms (Halstead and O'Rourke, 1977a, 1977b). Even though there have been intensive studies on the adaptive immune response to the dengue virus, the precise process that leads to DHF remains unclear. One of the missing links is the contribution of innate immune response, especially during the early phase of infection. Natural killer cells (NK cells) are key innate immune cells which provide prompt responses to pathogen and able to bridge between innate and adaptive immunity.
NK cells eliminate pathogens by several mechanisms. These include cytolysis via the perforin/granzyme pathway, triggering apoptosis through the interaction between Fas and Fas ligand, and the production of cytokines such as IFN-γ, TNF-α and MIP-1β. Additionally, NK cells can coordinate with antibodies to destroy virus infected cells through a process known as antibody dependent cell cytotoxicity (ADCC). NK cells can be divided into 2 groups according to their CD56 surface expression; CD56bright, which are potent cytokine producers and CD56dim, which are efficient at lysis of target cells. There are 3 main categories of NK receptors. The first category is Natural Cytotoxic Receptors (NCRs) consisting of NKp30, NKp44 and NKp46. These NCRs are activating receptors and recognize tumour-induced or pathogen-derived molecules. The second category is made up of lectin-like receptors (NKG2), which include NKG2A, NKG2C, and NKG2D. The NKG2 family of receptors can be either activating or inhibitory. NKG2A and NKG2C form heterodimers with CD94 proteins and bind to Human Leukocyte Antigen – E (HLA-E) molecules. NKG2D, on the other hand, exists as a homodimer and can recognize several ligands, including stress-inducible proteins such as MICA/B, ULBP (Thielens et al., 2012). The third category consists of Killer-cell Immunoglobulin-like Receptors (KIRs). These can be either inhibitory or activating receptor, depending on the immunoreceptor tyrosine-based motif present in their cytoplasmic tails. KIRs are capable of recognizing HLA class I molecules. For instance, KIR2DL1/L2/L3/S1 binds to allotypes of HLA-C, KIR3DL1/S1 targets HLA-Bw*4, and KIR3DL2/S2 recognizes HLA-A*3 and HLA-A*11 (Saunders et al., 2015). The activation or silence of NK cells are tightly regulated by a balance between signals from activating and inhibitory NK receptors expressed on the cell surfaces (Vivier et al., 2008).
The significance of NK cells has been shown in several virus infections. The lack of NK cells causes severe herpesvirus infection (Biron et al., 1989). Engagement of KIR3DS1+ NK cells and their cognate ligand, leads to NK activation and lysis of human immunodeficiency virus (HIV) infected cells which correlates well with slow progressor to AIDS (Alter et al., 2007). The selective expansion of CD94/NKG2C+KIR2DL2/L3+ NK cells with cytotoxicity function is associated with the control of acute chikungunya infection (Petitdemange et al., 2011). In dengue disease, previous studies have demonstrated the increased level of activated NK cells in dengue patients and the dengue E protein has been found to bind to NKp44 receptor suggesting the involvement of NK cells in dengue virus infection (Green et al., 1999; Hershkovitz et al., 2009). This study aimed to investigate the role of NK cells by examining the NK cell profiles in dengue infected patients and exploring the mechanism that drive NK activation. The insights gained from this study will enhance the understanding of the innate immune responses and their roles in the dengue disease.
2. Materials and methods
2.1. Patient samples
Blood samples were taken from patients admitted to the Pediatric Department of Khon Kaen and Songkhla hospitals, Thailand, after informed consent and approval from the ethical committees of Khon Kaen, Songkhla, Siriraj hospitals (Ref no. 92/2550) and Oxford University. Acute dengue infection was identified by RT-PCR-based gene identification or dengue-specific IgM capture ELISA (Innis et al., 1989; Yenchitsomanus et al., 1996). Secondary dengue infection was defined as a dengue-specific IgM/IgG ratio <1.8, by IgM and IgG capture ELISA using patient's sera.
Disease severity was classified according to the World Health Organization criteria 1997 (World Health Organization, 1997). PBMCs were isolated from patient's blood collected on fever day 5–7 (acute) and on 2-month after onset of fever (convalescent) by Ficoll-Hypaque density gradient centrifugation and cryopreserved for the subsequent studies. The viability of PBMCs was greater than 90 % by Trypan blue dye exclusion method. The details of patients are shown in Table 1.
Table 1.
Details of dengue infected patients in this study.
| Disease severity | Number of subjects | Type of infection | Male: Female | Average age in years (min, max) |
|---|---|---|---|---|
| Dengue fever (DF) | 41 | secondary | 20:21 | 10.4 (8,12) |
| Dengue hemorrhagic fever (DHF) | 41 | secondary | 19:22 | 10.0 (6,13) |
2.2. Antibodies
The following fluorescent conjugated anti-human monoclonal antibodies (mAbs) were utilized in this study: anti-CD3 (clone UCHT-1 and clone SK7), anti-CD56 (clone HCD56), anti-CD69 (clone FN50), anti-IFN-γ (clone 4S.B3) and anti-TNF-α (clone MAb11) mAbs were from Biolegend. Anti-NKp30 (clone Z25), anti-NKp44 (clone Z231), anti-NKG2A (clone Z199), anti-KIR2DL1/S1 (clone EB6B) and anti-KIR3DL1/S1 (clone Z27) mAbs were from Beckman Coulter. Anti-NKp46 (clone 9E2/NKp46), anti-KIR2DL2/L3/S2 (clone DX27), and anti-CD107a (clone H4A3) mAbs were from BD Bioscience. Anti-NKG2C (clone 134,591) mAb was from R&D system. Biotinylated anti-NKG2D (clone 1D11) mAb and APC conjugated streptavidin were from eBioscience.
2.3. Monocyte-derived dendritic cells (DCs) preparation and in vitro infection with dengue virus
PBMCs were isolated from healthy donors as previously described in patient samples. A fraction of cells was cryopreserved and subsequently used as effector. Autologous CD14+ monocytes were isolated from PBMCs by positive selection using CD14 Microbeads according to manufacturer's instruction (Miltenyi Biotec). Isolated monocytes were cultured for 5-6 days at 37 °C, 5 % CO2 incubator in RPMI1640 supplemented with 10 % FBS (R10), 20 ng/ml human rGM-CSF (First Link) and 25 ng/ml human rIL-4 (eBioscience) to differentiate into immature DCs. Phenotype of DCs were accessed by staining with CD14 and DC-SIGN antibodies. DCs (CD14−DC-SIGN+ cells) were infected with DENV2 strain 16,681 at multiplicities of infection (MOI) of 1 at 37 °C for 2 h. DCs treated with mock were used as negative control. After incubation, cells were washed with RPMI 1640, and plated at density of 5 × 105 cells/ml in R10 at 37 °C in 5 % CO2 incubator for overnight. All DCs-related processes had been performed under LPS-free conditions.
2.4. In vitro stimulation of NK cells
For cell stimulation with dengue-infected DCs, PBMCs were co-cultured with either mock-treated or DENV-infected autologous DCs at effector to target (E:T) ratio of 2:1 at 37 °C in 5 % CO2 incubator for 6 or 24 h. For 6 h co-culture, anti-human CD107a was added at the beginning of incubation, and the protein transport inhibitor, brefeldin A (BFA; Sigma; 10 µg/ml) and monensin (BD Bioscience; 0.7 µl/ml), were added after 1 h incubation. For 24 h co-culture, the mixture of anti-CD107a, BFA and monensin were added at the last 6 h of incubation. Activated PBMCs stimulated with 100 ng/ml Phorbol 12-myristate 13-acetate (PMA) and 500 ng/ml Ionomycin were used as positive control and resting PBMCs in R10 were used negative control. After that, cells were harvested, and immunofluorescence staining was performed.
For cell stimulation with dengue virus, PBMCs were co-cultured with DENV, ultraviolet (UV)-inactivated DENV at MOI of 1 or 10 for 6 and 24 h at 37 °C in 5 % CO2 incubator. Cells culture with mock supernatant or R10 were used as negative control.
2.5. Blocking experiment
PBMCs or DENV-infected DCs were treated with mAbs specific to IL-2Rα, IL-12, IL-15 (R&D system), IL-18 (MBL), type I IFNR (PBL) for 1 h prior to co-culture steps. Cells treated with isotype matched controls (R&D system) were used as negative control. The final concentration of each Ab was 10 µg/ml.
2.6. Transwell experiment
Mock-treated or DENV-infected DCs (5 × 105 cells) were seeded on the lower chamber of 24-well transwell plate (Transwell permeable support 0.4 μm; Corning) and incubated at 37 °C in 5 % CO2 incubator for overnight. The next day, autologous PBMCs (1 × 106 cells) were plated on the upper chamber of the transwell plate, and incubated further for 6 or 24 h. The mixture of PBMCs and mock-treated or DENV-infected DCs was used as controls. After the incubation, PBMCs were harvested, and immunofluorescence staining was performed.
2.7. FACS analysis
For the phenotypic analysis, healthy donors, or patients’ PBMCs were blocked for Fc receptors by the combination of 10 % of human AB serum and 10 % of mouse serum at 4 °C for 10 min followed by surface staining with panels of mAbs at 4 °C for 30 min. Next, cells were washed with FACS wash (1 % FBS 0.5 % human serum, 2 mM EDTA, 0.1 % NaN3 in PBS), fixed in FACS fix (1 % formaldehyde in PBS) and analyzed by flow cytometer.
For the functional analysis, after surface staining, cells were fixed with 4 % formaldehyde and permeabilized with 0.5 % saponin. Permeabilized cells were stained with anti-IFN-γ and/or anti-TNF-α mAbs and incubated further at 4 °C for 30 min. Thereafter, cells were washed, fixed in FACS fix and analyzed by flow cytometer. Data was analyzed by using FlowJo software (version 10.1; TreeStar).
2.8. Statistical analysis
The statistical analysis was performed using the Prism program (Graph Pad Software). Differences between ex vivo acute and convalescent NK cells phenotypes in DF or DHF groups, kinetics of NK cells responses at 6 and 24 h in vitro co-culture, the expression of NK receptors on IFN-γ+ and IFN-γ− NK cells were all evaluated by nonparametric paired t-test (Wilcoxon signed rank test). Differences between DF-DHF groups were analyzed using nonparametric unpaired t-test (Mann-Whitney U test). The value of p ≤ 0.05 was statistically significant.
3. Results
3.1. Proportional alteration and activation of CD56bright and CD56dim NK cells during acute dengue disease
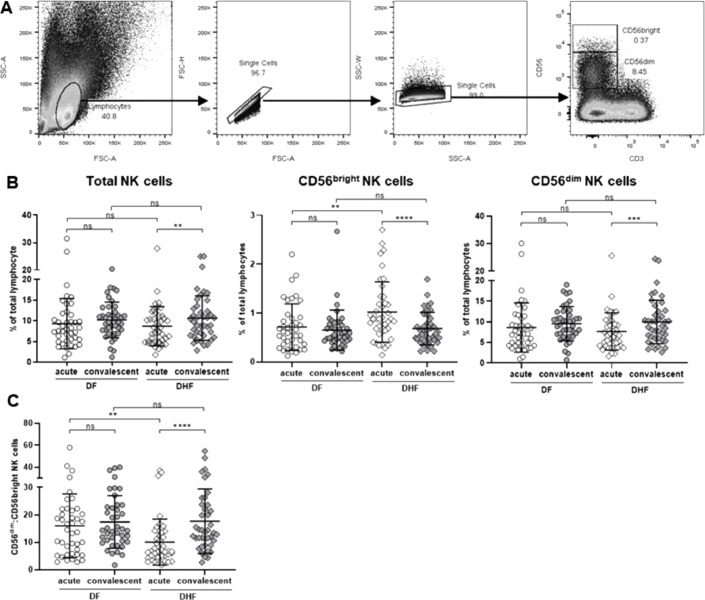
Profiles of NK cells obtained from dengue patients (DF and DHF groups) at two time points; the time when the symptoms occur (acute) and the time when patients were fully recovered (2-month convalescent), were evaluated. PBMCs were stained with a set of monoclonal antibodies specific to NK markers and activation status. NK cells were defined as CD56+CD3− population (Fig. 1, Fig. 2). The gating strategies are shown in Fig. 1A.
Fig. 1.
Frequencies of NK cells in dengue infected patients during acute and convalescent periods
(A) Lymphocytes were gated based on FSC-A vs. SSC-A, and singlet cells were further gated from FSC-A vs. FSC-W. NK cells were defined as CD56+CD3− cells, and their subpopulations, CD56bright and CD56dim, were gated based on the levels of CD56 expression. (B) Scattered plots demonstrated the frequencies of NK cells; total NK cells and subsets of NK cells (CD56bright and CD56dim NK cells) (C) The ratio between frequencies of CD56dim and CD56bright NK cells in DF and DHF patients. Horizontal bars indicate the mean and error bars indicate standard deviation (SD). Differences between acute and convalescent samples and between DF and DHF were analyzed using Wilcoxon signed rank test and Mann-Whitney U test, respectively. Asterisks indicate P-values: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. “ns.” indicates a non-statistically significant.
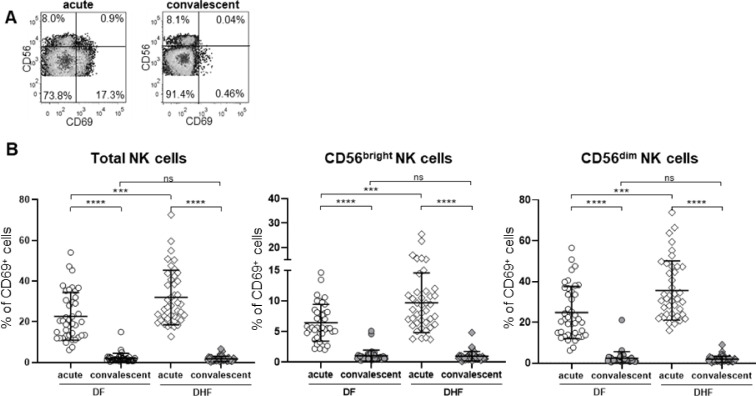
Fig. 2.
Activation of NK cells in dengue infected patients’ PBMCs ex vivo.
(A) Representative's plots of CD69 expression on NK cell subpopulations of acute and 2-month convalescent samples. (B) The frequencies of CD69+CD56+ cells (left), CD69+CD56bright cells (middle) and CD69+CD56dim NK cells (right). The frequencies of NK cells that express CD69, an activation marker, were determined. Data are shown as scattered plots of acute and 2-month convalescent samples from DF and DHF patients. Horizontal bars indicate the mean and error bars indicate standard deviation (SD). Differences between acute and convalescent samples were determined using Wilcoxon signed rank test. Differences between DF and DHF samples were determined using Mann-Whitney U test. Asterisks indicate P-values: *P<0.05, **P<0.01, ***P<0.001, ****P < 0.0001. “ns.” indicates a non-statistically significant.
Frequencies of NK cells were similar between DF and DHF either at acute or convalescent. However, DHF patients showed the reduction of NK cells during acute period comparing to that of convalescent (8.7 ± 4.8% vs. 10.6 ± 5.4 %; p = 0.004) whilst levels of NK cells in DF patients remained unchanged over time (9.3 ± 6.1 % vs. 10.2 ± 4.3 %, p = 0.32) (Fig. 1B, left). NK cells can be divided into 2 subsets (CD56bright vs. CD56dim NK cells). The alterations of NK cells subpopulations were further investigated. Interestingly, in acutely infected DHF patients, despite the lower number of total NK cells, an expansion of CD56bright NK cells was observed with the higher level comparing to convalescent period (acute vs. convalescent, 1.02 ± 0.62 % vs. 0.68 ± 0.33 %; p < 0.0001; Fig. 1B, middle). Meanwhile, the pattern of CD56dim NK cells followed the same trend as total NK cells in which the lower frequencies of cells were found in acute DHF comparing to convalescent (7.7 ± 4.5 % vs. 10 ± 5.3 %, p = 0.001; Fig. 1B, right). The ratio between CD56dim and CD56bright NK cells were significantly lower in acute DHF compared to convalescent phase and acute DF (acute vs. convalescent, 10.0 ± 8.4 vs. 17.7 ± 11.8, p < 0.0001; Fig. 1C). Such alteration was clearly seen either by frequencies or absolute number of NK cells (data not shown). Unlike DHF, frequencies of CD56bright and CD56dim NK cells derived from DF patients were similar at both points of time. Interestingly, we observed that DHF patients contained a significantly higher number of CD56bright NK cells than DF during acute phase of infection (acute DF vs. acute DHF, 0.71 ± 0.48 % vs. 1.02 ± 0.62 %, p = 0.007; Fig. 1B, middle).
Next, the activation status of NK cells was accessed based on CD69 surface protein expression. The representative FACS profile is shown in Fig. 2A. The elevation of activated NK cells at acute phase were detected in both DF (acute vs. convalescent, 22.7 ± 11.7 % vs. 2.1 ± 2.4 %; p < 0.0001) and DHF patients (acute vs. convalescent, 32.0 ± 13.4 % vs. 1.8 ± 1.3 %, p < 0.0001; Fig. 2B, left). Notably, the levels of activated NK cells were correlated with disease severity (acute, DF vs. DHF, 22.7 ± 11.7 % vs. 32.0 ± 13.4 %, p = 0.0006). Similar patterns were observed in both subsets of NK cells in patients with either DF or DHF as shown in Fig. 2B, middle and right.
In conclusion, NK cells were activated during acute phase of disease in both DF and DHF patients. However, NK cells from DHF patients showed some characters that differed from DF. During acute period, despite being highly activated, the total frequency of NK cells from DHF patients decreased and the subset was skewed toward the increase of CD56bright NK populations.
3.2. Alteration of activated NK cell phenotypes: the more up-regulation of activating receptors in mild dengue disease
Functions of NK cells are strictly controlled by the signals from receptors presented on the cell surfaces, activating and inhibitory receptors. The profiles of both activating and inhibitory NK receptors in dengue infected patients were examined. Any receptors that could discriminate DHF from DF patients were of interest. NK receptors included in this study were natural cytotoxic receptors (NKp30, NKp44, and NKp46), lectin-like receptors (NKG2A, NKG2C and NKG2D) and killer immunoglobulin-like receptor (KIR2DL1/S1, KIR2DL2/L3/S2 and KIR3DL1/S1). The frequencies of individual receptors were compared between activated (CD69+) and non-activated (CD69−) NK cells collecting during acute and 2-month convalescent periods, respectively. The details of the frequencies of these receptors in CD56bright and CD56dim NK cells subpopulations are shown in Supplementary Figs. 1 and 2, respectively.
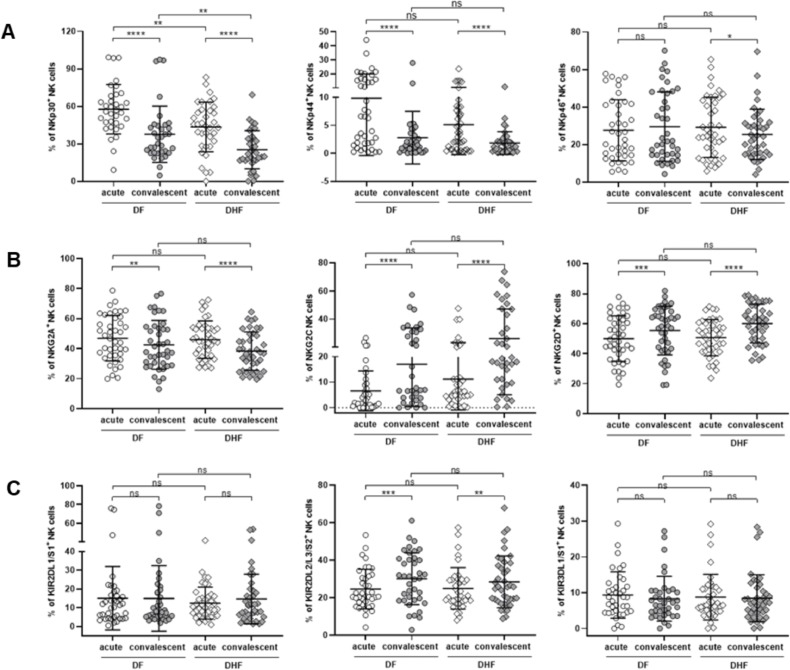
For natural cytotoxic receptors, the expression level of NKp30 in activated NK cells was significantly increase during acute phase of infection in both DF (57.6 ± 19.7 % vs. 37.7 ± 22.5 %, p < 0.0001) and DHF (43.5 ± 19.9 % vs. 25.3 ± 15.3 %, p < 0.0001). Furthermore, the expression of NKp30 in activated NK cells was higher in DF patients compared to DHF at both points of time (acute, DF vs. DHF, p = 0.006; convalescent, DF vs. DHF, p = 0.009, Fig. 3A, left). A similar pattern was observed in NKp44 expressing cells. NKp44, an activating NK receptor which has been shown to interact with the dengue E protein, was upregulated in activated NK cells during acute phase in both DF and DHF groups and there was a trend that levels of NKp44+CD69+ NK cells in DF patients were higher than that of DHF although the differences could not reach statistical significance (9.8 ± 10.2 % vs. 5.1 ± 5.4 %, p = 0.07; Fig. 3A, middle). Unlike NKp30 and NKp44 receptors, the frequencies of NKp46 expressing cells were comparable between DF and DHF patients. However, levels of NKp46+ NK cells were selectively increase in acute DHF, not in acute DF, compared to convalescent (acute vs. convalescent: DHF; 29.2 ± 16.1 % vs. 25.4 ± 13.4 %, p = 0.02, DF; 27.6 ± 16.3 % vs. 29.5 ± 18.5 %, p = 0.19; Fig. 3A, right). Strikingly, NKp30 is the only NK receptor that could discriminate between DF and DHF patients.
Fig. 3.
Profiles of NK receptors on NK cells from dengue infected patients
Scattered plots of frequencies of NK cells expressing indicated NK receptors in DF and DHF patients are demonstrated. Expression of individual NK receptors were determined in activated NK cells (CD56+CD3−CD69+) in acute samples and in non-activated NK cells (CD56+CD3−CD69−) in convalescent samples. (A) Natural cytotoxic receptors (NCRs): NKp30 (left), NKp44 (middle) and NKp46 (right). (B) Lectin-like receptors (NKG2- family): NKG2A (left), NKG2C (middle) and NKG2D (right). (C) Killer immunoglobin-like receptors (KIRs): KIR2DL1/S1 (left), KIR2DL2/L3/S2 (middle) and KIR3DL1/S1 (right). Horizontal bars indicate means, and the error bars indicate standard deviation (SD). Differences between acute and convalescent samples were determined using Wilcoxon signed rank test. Differences between DF and DHF samples were determined using Mann-Whitney U test. Asterisks indicate P-values: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. “ns.” indicates a non-statistically significant.
In contrast to the NCRs, there was no difference in expression pattern of the NKG2 family receptors between DF and DHF groups. However, some changes between NK cells collected during acute and at 2-month convalescent were observed. NKG2A and NKG2C receptors recognize the same HLA-E molecules. However, interaction with NKG2A leads to the inhibition of NK cells, while engagement with NKG2C results in NK cells activation. the Frequencies of NKG2A expressing cells were higher at acute phase compared to that of convalescent in both DF (acute vs. convalescent; 46.9 ± 15.2 % vs. 42.5 ± 16.2 %; p = 0.006) and DHF patients (acute vs. convalescent; 46.0 ± 12.5 % vs. 38.3 ± 12.8 %; p < 0.0001) (Fig. 3B, left). On the other hand, levels of NKG2C expressing cells were lower during acute period in DF compared to that of convalescent (acute vs. convalescent; 6.6 ± 7.8 % vs. 17.1 ± 16.5 %; p < 0.0001) and the same trend was found in DHF patients (acute vs. convalescent; 11.2 ± 12.1 % vs. 26.1 ± 21.0 %; p < 0.0001) (Fig. 3B, middle). Expression pattern of NKG2D, another activating NK receptor, followed the same trend as NKG2C. Frequencies of NKG2D positive cells was lower during acute phase, in both DF (acute vs. convalescent; 50.0 ± 15.2 % vs. 55.4 ± 16.2 %; p = 0.001) and DHF (acute vs. convalescent; 50.8 ± 12.1 % vs. 60.1 ± 12.9 %; p < 0.0001) (Fig. 3B, right). Additional work is required to clarify the implications of the inversed correlations between inhibitory and activation NKG2 receptors.
Among three types of KIRs, KIR2DL2/L3/S2 were the only receptors that altered during the course of infection. The frequencies of KIR2DL2/L3/S2 expressing NK cells were lower during acute phase in both DF (acute vs. convalescent, 24.6 ± 10.6 % vs. 30.1 ± 18.3 %; p = 0.001) and DHF patients (acute vs. convalescent, 24.9 ± 11.1 % vs. 28.5 ± 13.8 %; p = 0.004; Fig. 3C, middle). Ligands of KIR2DL2/L3/S2 are HLA-C allotypes. Meanwhile, the expression of KIR2DL1/S1 which also binds to HLA-C molecule, did not change over time (Fig. 3C, left). Similar pattern was observed in KIR3DL1/S1+ NK cells which recognize HLA-Bw4 alleles (Fig. 3C, right). In line with NKG2 family receptors, none of the KIRs could discriminate DF from DHF. It should be noted at the time when we conducted the experiment, there was no commercially available antibody that could discriminate between activating and inhibitory forms of KIRs due to the high homology of the ectodomain. Thus, there was some limitation of the precise phenotyping of KIRs.
Taken together, these findings demonstrate the dynamic of NK receptor expressions in activated NK cells during acute periods. Such alterations reflect the contribution of NK cells in immune responses to dengue virus. Importantly, the NKp30, an activating NK receptor, was the only receptor that could distinguish between DF and DHF.
3.3. Co-cultured with DENV infected DCs could elicit NK cells responses and IFN-γ production and degranulation were the key functions
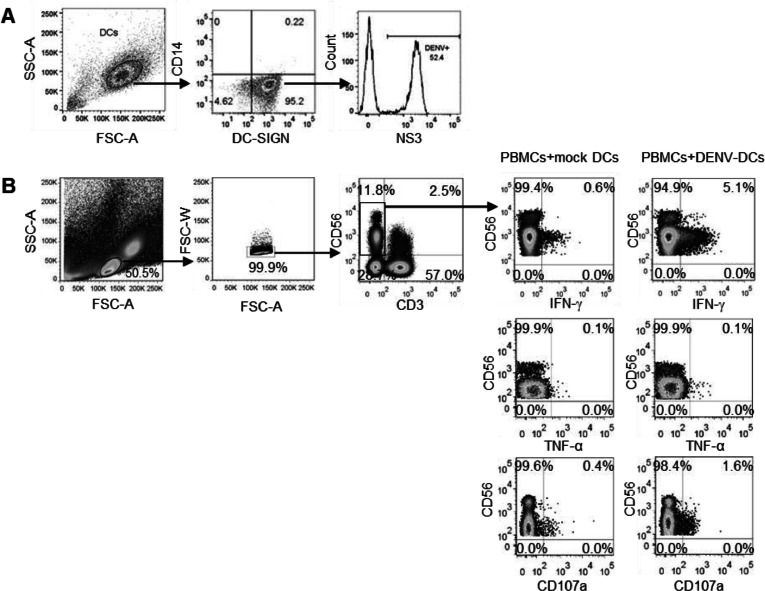
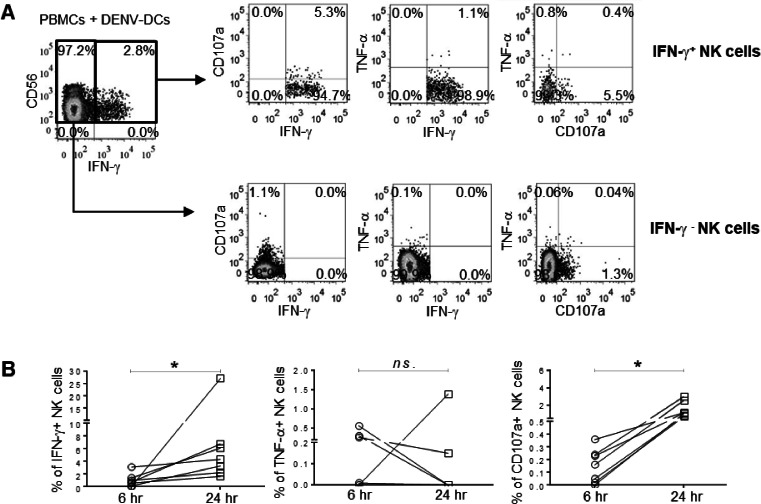
The ex vivo analysis in dengue infected patients revealed that NK cells were activated and exhibited the changes of NK receptors during acute period, it would be interesting to determine the mechanism by which NK cells respond to the dengue virus. As our cohort consists of pediatric dengue patients, the blood volume was limited. We decided to set up the in vitro experiments using PBMCs from healthy donors to dissect the mechanism of NK cells. The functional NK cells were accessed by detection of CD107a (marker of degranulation which reflects cell cytolysis), IFN-γ and/or TNF-α expressing NK cells. To explore whether dengue virus could directly trigger NK cells responses, PBMCs isolated from healthy donors were cultured with dengue viruses or UV-inactivated viruses in a presence of protein transport inhibitors. However, we could not detect IFN-γ producing NK cells even at 24 h post stimulation with high amount of virus (MOI of 10), indicating that the virus alone could not activated the NK cells (Supplementary Fig. 3). Since crosstalk between NK cells and dendritic cells (DCs) have been reported and DCs are the major target of dengue virus (Tassaneetrithep et al., 2003). Autologous monocytes derived DCs were infected with dengue virus and used as target cells in the in vitro NK cells stimulation. The frequencies of dengue infected DCs were determined by staining with dengue NS3 protein-specific antibodies as shown in Fig. 4A. Through this system, we were able to activate NK cells as shown by the expression of CD107a or the production of IFN-γ by NK cells at 24 h post-stimulation (Fig. 4B). The kinetic assay was then conducted to determine whether the activation of NK cells by DENV infected DCs were time dependent. Two cell stimulation periods were performed: one at 6 h and another at 24 h. These periods represent the early and late responses, respectively. In these experiments, we also explored the TNF-α production of NK cells upon activation. The representative FACS profiles is shown in Fig. 5A. IFN-γ producing NK cells could be detected early at 6 h of cell stimulation and levels were statistically significantly increase at 24 h (6 vs. 24 h, 1.04 ± 1.05 % vs. 7.40 ± 8.95 %; p = 0.02) (Fig. 5B, left). CD107a expressing NK cells followed the same trend with albeit lower number of responding cells (6 vs. 24 h, 0.15 ± 0.13 % vs. 1.51 ± 0.89 %; p = 0.02, Fig. 5B, right). Thus, the responses of NK cells were increased over time. Interestingly, the patterns of functional NK cells were monotypic i.e., IFN-γ+ only or CD107a+ only. There were a few cells that produced both IFN-γ and CD107a (Fig. 5A). To our surprise, the TNF-α producing NK cells were very low and at any point of times (Fig. 5B, middle). Of note, NK cells were able to secrete TNF-α upon stimulation with PMA and ionomycin, indicating that these cells were functional (data not shown).
Fig. 4.
Stimulation of NK cells by DENV-infected DCs.
DCs were treated with mock or infected with DENV (MOI 1) for overnight, and the infection was detected intracellularly by anti-dengue antibodies. (A) The representative FACS profile of dengue infected DCs. Cells were gated based on FSC-A and SSC-A, followed by CD14 and DC-SIGN expression. Dendritic cells (CD14−DC-SIGN+) were gated and analyzed for DENV NS3 antigen expression. Histogram demonstrates frequencies of DENV infected DCs as shown by positive staining with dengue NS3 protein specific mAbs. (B) The representative FACS profile of NK cells (CD56+CD3−) upon stimulation with mock or dengue infected DCs for 24 h. The frequencies of IFN-γ+, TNF-α+ and CD107a+ NK cells are demonstrated.
Fig. 5.
Kinetics of NK cells responses against DENV-infected autologous DCs.
(A) The representative plots show the pattern of NK cells responses against DENV-infected DCs at 24 h. (B) The summary of frequencies of IFN-γ+ (left), TNF-α+ (middle) or CD107a+ (right) NK cells at 6- and 24 h co-culture. The differences of NK cells responses between two time points were analyzed using Wilcoxon signed rank test. Asterisks indicated P-values: *P<0.05. “ns.” indicates a non-statistically significant.
3.4. The mechanisms of NK stimulation by DENV-infected DCs: both cytokines and cell-cell contact dependent
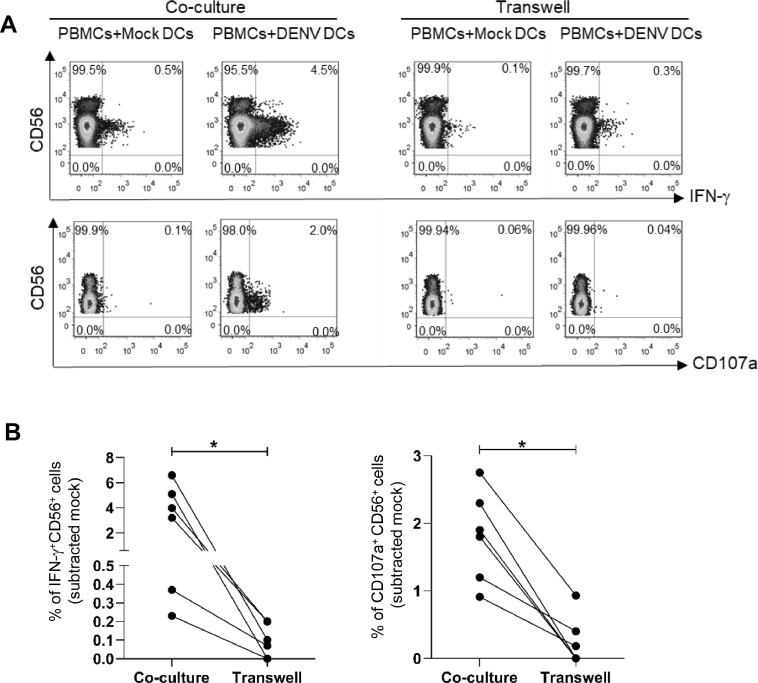
Since NK cells can be activated through engagement of NK cell receptors and cytokine signaling (Haeberlein et al., 2010; Madera et al., 2016). We sought to know whether the response of NK cells to dengue viruses occur by which mechanisms. PBMCs were co-cultured with autologous DENV infected DCs in 2 conditions: within the same well or within the same well but in different compartments (transwell). As shown in Fig. 6A, IFN-γ production or degranulation of NK cells could be observed when both effectors and target cells were incubated in the same compartment, but the responses were significantly decreased in transwell condition (Fig. 6B) indicating that direct contact with dengue infected cells is important for NK activation.
Fig. 6.
Direct contact between NK cells and DENV-DCs elicit NK cells activation.
(A) The representative plots show the frequencies of IFN-γ+NK cells (upper row) and CD107a+ NK cells (lower row) from PBMCs-DENV-infected autologous DCs co-culture with or without 0.4 µm transwell at 24 h. (B) Line graphs demonstrate the frequencies of IFN-γ+NK cells (left) and CD107a+ NK cells (right) in co-culture and transwell conditions from 6-independent experiments. Asterisks indicated P-values: *P<0.05.
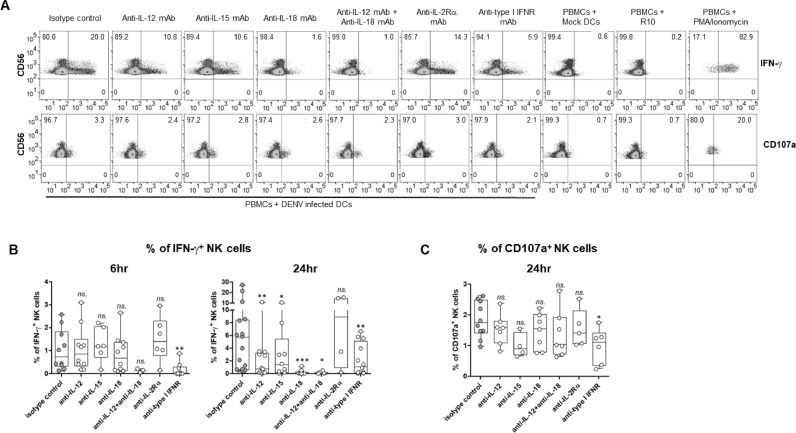
In addition, previous studies from our group and others, demonstrated that DENV infected DCs produced several cytokines that may have an impact on NK function such as IL-12p40, IL-18, type I IFN, and IFN-γ (Dejnirattisai et al., 2008; van Wilgenburg et al., 2016). To investigate the role of cytokines on NK cell responses, PBMCs were stimulated with DENV-infected DCs in the presence of blocking antibodies (monoclonal antibodies specific to IL-12, IL-15, IL-18, IL-2 receptor, and type I IFN receptor). The blocking effect was accessed at two periods, 6 h and 24 h post cell stimulation. Indeed, blockade of certain cytokines led to a reduction in IFN-γ production or CD107a expression by NK cells. Furthermore, the blocking effect of individual cytokines varied over time, indicating the kinetics of NK cell responses to dengue virus. The representative FACS profiles are depicted in Fig. 7A.
Fig. 7.
Effect of cytokines on NK cells activation in responding to dengue infected DCs.
PBMCs and DENV-infected autologous DCs were co-cultured with 10 µg/ml of indicated blocking mAbs or isotype-matched control (n = 4–12) for 6 or 24 h. (A) Representative FACS plots demonstrate frequencies of IFN-γ (top) or CD107a (bottom) positive NK cells (CD56+CD3−) in a presence of blocking mAbs for 24 h. PBMCs co-cultured with mock-treated DCs or R10 and PMA/Ionomycin were used as negative and positive control, respectively. (B) Box and whisker plots demonstrate the frequencies of IFN-γ+ NK at 6 h (left) and 24 h (right) stimulation. (C) Box and whisker plots demonstrate the frequencies of CD107a+ NK cells at 24 h stimulation. The differences of NK cells responses comparing to isotype control were analyzed by Wilcoxon matched-pairs signed rank test. Asterisks indicated P-values: *P < 0.05, **P < 0.01, ***P < 0.001 and “ns.” indicates a non-statistically significant.
At 6 h incubation, only blocking of type I IFN receptor (anti-type I IFNR) significantly reduced frequencies of IFN-γ secreting NK cells whilst blocking with other cytokines had no effect (Fig. 7B left). Therefore, IFN-γ production of NK cells during the early phase of responses is involving with type I IFNs signaling. Because of the low frequencies of CD107a expressing NK cells at 6 h stimulation (as shown in Fig. 5B), we did not determine the blocking effect to CD107a expression at this time point. At 24 h incubation, the presence of all blocking antibodies, except for the anti-IL-2 receptor mAb, led to a decrease in the frequencies of IFN-γ producing NK cells (Fig. 7B right). The most significant blocking effect was observed with IL-18 mAb, followed by IL-12, IL-15, and a combination of IL-12 and IL-18 mAbs. The impact of type I IFN was slightly decreased as compared to 6 h stimulation (% of reduction, mean ± SD: 90.82 ± 13.77 % at 6 h vs. 51.43 ± 33.59 % at 24 h). In terms of degranulation, only blocking of type I IFN significantly decreased the frequencies of CD107a expressing NK cells (Fig. 7C). These results indicate the importance of cytokines in eliciting responses of NK cells. IL-12, IL-15 and IL-18 significantly involved in the production of IFN-γ. Meanwhile, type I IFNs play a key role in both degranulation and the cytokine production of NK cells.
Taken together, these results demonstrated that dengue virus infected DCs triggers NK cell responses via cell-cell contact and cytokine signaling.
4. Discussion
In this study, the frequencies, and phenotypes of peripheral blood NK cells of patients with dengue fever (DF) and dengue hemorrhagic fever (DHF) were followed from the day of onset fever to the recovery phase. We found a significant activation in both subsets of NK cells (CD56bright and CD56dim), with the degree followed disease severity. This finding highlights the importance of NK cells in dengue pathogenesis which in agreement with the previous published reports (Green et al., 1999; Keawvichit et al., 2018; Zimmer et al., 2019). In DHF patients, despite the highly activated phenotype, the total frequencies of NK cells were reduced compared to those observed during the convalescent period. There was a skewed repertoire of NK subsets as the levels of CD56bright NK cells increased whilst the CD56dim NK cells decreased. Consequently, the ratio between CD56dim and CD56bright NK cell subsets was significantly lower. One of the factors driving the expansion of CD56bright NK population is type I IFNs. High levels of IFN-α have been detected in serum of dengue patients especially in DHF, 2 or 3 days before defervescence (Kurane et al., 1993; Dejnirattisai et al., 2008). Thus, it is plausible that the presence of IFN-α in serum might stimulate the proliferation of CD56bright NK cells population leading to the lower ratio of CD56dim/bright NK cells. However, other explanation that the reduced ratio is from the decreased number of CD56dim population or the defect in maturation process changing from CD56bright to CD56dim population are still cannot be excluded. The expansion of CD56bright NK cells have been reported in several virus infections in human such as in hepatitis C (HCV) patients who received IFN-α and ribavirin treatment (Lee et al., 2010), HIV-1 infected patients (Jiao et al., 2015), HCV and HIV coinfected patients (Bhardwaj et al., 2016) and systemic lupus erythematosus (SLE) patients (Schepis et al., 2009). Vaccination with adenovirus expressing Zaire Ebola virus (ZEBOV) glycoprotein and modified vaccinia Ankara expressing ZEBOV, Sudan Ebola and Marburg virus GPs and Tai Forest Ebola virus nucleoprotein (MVA-BN-Filo) also induced a significant increase of CD56bright NK cells (Wagstaffe et al., 2020). As CD56bright NK cells are potent cytokine producers. We speculated that the cytokines secreted from CD56bright NK cells might be one of the factors contributing to cytokines storms in DHF patients. Importantly, apart from activation status, there were alterations of cell surface receptors in activated NK cells and these changes could be observed in all NK receptors family.
Both NKp30 and NKp44, natural cytotoxic receptors, were elevated during acute period in both DF and DHF patients and NKp46 expressing cells were higher in DHF patients. Since all NCRs were upregulated in activated NK cells, understanding of dengue components that specifically bind to these receptors is crucial. Previous study demonstrated the binding between soluble dengue E protein and the recombinant NKp44 but not the NKp30 nor NKp46D2 Ig fusion proteins suggesting that the E protein is a ligand of NKp44 receptor (Hershkovitz et al., 2009). Concurrently, the ligands for NKp30 and NKp46 which related to the dengue virus have not been identified. In other diseases, NKp30 receptor binds to B7H6 and heparan sulfate proteoglycans (HSPGs), endogenous ligands expressed on tumor cells (Bloushtain et al., 2004; Matta et al., 2013) and pp65 protein of human CMV (Arnon et al., 2005). NKp46 receptor binds to heparan sulfate (HS) glycosaminogylcans (GAGs), haemagglutinin of influenza virus and the haemagglutinin-neuraminidase of parainfluenza virus (Hecht et al., 2009; Mandelboim et al., 2001). Future work is needed to determine the ligands of these receptors in the context of dengue virus.
Apart from natural cytotoxic receptors, NKG2A receptor (inhibitory receptor) was upregulated whilst NKG2C and NKG2D receptors (activation receptor) were downregulated in activated NK cells. Previous studies demonstrated the elevation of corresponding ligands to these receptors in acutely infected dengue patients. The increase level of HLA-E molecules, the ligand of NKG2A and NKG2C receptors (McKechnie et al., 2019) and the association between MICA and MICB molecules, ligands of NKG2D receptor with symptomatic dengue infection, have been reported (Garcia et al., 2011; Luangtrakool et al., 2020). MICA*045 and MICB*004 alleles are associated with susceptibility to DHF whilst MICB*002 alleles are associated with protection from DHF in secondary dengue infection (Garcia et al., 2011; Luangtrakool et al., 2020). Given the increased levels of inhibitory NKG2 receptors but not the activating receptors, we speculate that the NKG2 receptors might play role in dampening NK cell responses. Downregulation of KIR2DL2/L3/S2 in activated NK cells was observed in both DF and DHF patients. Due to the limitations of available antibodies, we could not dissect whether the reduced population was from cells expressing activation (KIR2DS2) or inhibitory receptors (KIR2DL2 and 2DL3). Nevertheless, previous study has shown that KIR2DS2 expressing NK cell line recognizes the conserved dengue NS3 peptide presented on HLA-C*0102 molecule and resulting in cytolysis of target cells (Naiyer et al., 2017). Genetic study showed that the frequencies of KIR2DS2 in dengue-infected patients were lower compared to healthy controls (Petitdemange et al., 2014). Meanwhile, expression of KIR2DL2 gene was associated with mild dengue disease (Tapsoba et al., 2022). Although we could not detect the changes of KIR3DL1/S1 receptor at any point of time, the binding between KIR3DL1 and dengue NS1 peptide-HLA-B*57 tetrameric complexes have been reported. These NS1 tetramer binding NK cells were markedly activated in severe dengue patients during febrile phase (Townsley et al., 2016). Collectively, these findings suggest the potential significance of NK receptors in dengue diseases. However, additional work is needed to fully understand the contribution of these receptors in dengue pathogenesis.
Importantly, the levels of NKp30 expressing activated NK cells were statistically significantly higher in DF patients compared to DHF. In line with our work, Shabrish et al. (2020) also observed increased frequencies of NKp30+ NK cells in adult patients with acute dengue fever. However, they could not detect the up regulation of NKp30+ NK cells in severe dengue (SD) patients and the levels were comparable to healthy controls group. Since our study specifically focused on pediatric dengue patients, rather than adults. The discrepancy could be attributed to variations in age groups as the pattern of NK cell receptors differs slightly across various ages (Almeida-Oliveira et al., 2011). Engagement between NKp30 and ligands not only triggers target cells cytolysis as has previously been shown in HIV, HCV and vaccinia virus infection (De Maria et al., 2003; Golden-Mason et al., 2010; Chisholm and Reyburn, 2006), but also promotes DC maturation and lysis of immature DCs (Ferlazzo et al., 2002). However, bindings of NKp30 with ligands could result in inhibition of NK cells as reported in human CMV infection and is believed to be one of the mechanisms that viruses employ to evade NK cell responses (Arnon et al., 2005). Therefore, it is important to explore the outcome of binding between NKp30 receptor and specific ligands in the context of dengue virus and its impact on dengue disease. To our knowledge, NKp30 is the first NK receptor that can distinguish between DF and DHF pediatric patients.
We and others demonstrated that NK cells activation could be elicited by direct interaction between dengue infected dendritic cells and NK cells (Costa et al., 2017; Lim et al., 2014). The binding was involved not only engagement of NK receptors but also several adhesion molecules such as 2β4, LFA-1, DNAM-1, and CD2 (Costa et al., 2017). Type I IFNs, IL-12, IL-15 and IL-18 secreted by dengue infected DCs also trigger NK cells responses, especially IFN-γ production. The elevated levels of these cytokines have also been detected in serum of dengue infected patients (Dejnirattisai et al., 2008; Zimmer et al., 2019; De et al., 2014; Mustafa et al., 2001; Pacsa et al., 2000). Importantly, the responses of NK cells were increased over time and exhibited a sequential effector function. The initial responses were IFN-γ production, followed by degranulation. Despite the ability of NK cells to execute multiple effector functions, the majority of responding cells showed only one function. This might be due to the different intracellular signaling pathway, IFN-αβ/STAT1 for cytotoxicity and IL-12/STAT4 for IFN-γ expression (Nguyen et al., 2002). Similar patterns have been observed by Zimmer et al. (2019) but in their study, NK cells from dengue patients were stimulated with HLA-Class I deficient cell lines and cytokines. The early IFN-γ production may influence the subsequent immune responses. It can drive DCs to secrete more cytokine such as IL-12 or chemokine such as IP-10 (Dejnirattisai et al., 2008; Libraty et al., 2001). In vitro studies demonstrated that IP-10 produced from DENV-DCs act as chemoattractant to recruit CXCR3+NK cells and CXCR3+T cells to the site of infection which resulting in amplification of immune responses (Dejnirattisai et al., 2008; Keawvichit et al., 2018).
In summary, our research emphasizes the complicated nature of NK cell responses to dengue virus infection. The marked activation of NK cells, coupled with alterations in specific NK receptors, a bias toward cytokine-producing NK cells, and the crosstalk with DCs, significantly influences the overall outcome of NK cell responses. Notably, NKp30 is the only receptor capable of distinguishing between DF and DHF in pediatric patients and could be potentially used to predict a mild outcome in individuals who are acutely infected with the dengue virus.
CRediT authorship contribution statement
Napas Taechasan: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis. Iris Scherwitzl: Methodology, Investigation, Formal analysis. Piyada Supasa: Methodology, Investigation, Formal analysis. Wanwisa Dejnirattisai: Methodology, Investigation, Formal analysis. Kanokwan Sriruksa: Resources. Wannee Limpitikul: Resources. Prida Malasit: Resources. Gavin R Screaton: Validation, Supervision, Methodology, Funding acquisition, Conceptualization. Juthathip Mongkolsapaya: Writing – review & editing, Validation, Supervision, Methodology, Funding acquisition, Conceptualization. Thaneeya Duangchinda: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Wellcome Trust Senior Investigation award (to G.R.S.), Mahidol Medical Scholars Program (to N.T.), the Research Excellence Development (RED) program, Faculty of Medicine Siriraj Hospital, Mahidol University (to W.D.) and National Science and Technology Development Agency (P-21-50031) (to T.D.). We thank Nattaya Tangthawornchaikul for collecting specimens.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2024.199382.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Almeida-Oliveira A., Smith-Carvalho M., Porto L.C., Cardoso-Oliveira J., Ribeiro Ados S., Falcao R.R., et al. Age-related changes in natural killer cell receptors from childhood through old age. Hum. Immunol. 2011;72(4):319–329. doi: 10.1016/j.humimm.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Alter G., Martin M.P., Teigen N., Carr W.H., Suscovich T.J., Schneidewind A., et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2007;204(12):3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon T.I., Achdout H., Levi O., Markel G., Saleh N., Katz G., et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 2005;6(5):515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- Bhardwaj S., Ahmad F., Wedemeyer H., Cornberg M., Schulze Zur Wiesch J., van Lunzen J., et al. Increased CD56(bright) NK cells in HIV-HCV co-infection and HCV mono-infection are associated with distinctive alterations of their phenotype. Virol. J. 2016;13:67. doi: 10.1186/s12985-016-0507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron C.A., Byron K.S., Sullivan J.L. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 1989;320(26):1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- Bloushtain N., Qimron U., Bar-Ilan A., Hershkovitz O., Gazit R., Fima E., et al. Membrane-associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. J. Immunol. 2004;173(4):2392–2401. doi: 10.4049/jimmunol.173.4.2392. [DOI] [PubMed] [Google Scholar]
- Bozza F.A., Cruz O.G., Zagne S.M., Azeredo E.L., Nogueira R.M., Assis E.F., et al. Multiplex cytokine profile from dengue patients: mIP-1beta and IFN-gamma as predictive factors for severity. BMC Infect. Dis. 2008;8:86. doi: 10.1186/1471-2334-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.E., Reyburn H.T. Recognition of vaccinia virus-infected cells by human natural killer cells depends on natural cytotoxicity receptors. J. Virol. 2006;80(5):2225–2233. doi: 10.1128/JVI.80.5.2225-2233.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V.V., Ye W., Chen Q., Teixeira M.M., Preiser P., Ooi E.E., et al. Dengue virus-infected dendritic cells, but not monocytes, activate natural killer cells through a contact-dependent mechanism involving adhesion molecules. mBio. 2017;8(4) doi: 10.1128/mBio.00741-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz Hernandez S.I., Puerta-Guardo H., Flores-Aguilar H., Gonzalez-Mateos S., Lopez-Martinez I., Ortiz-Navarrete V., et al. A strong interferon response correlates with a milder dengue clinical condition. J. Clin. Virol. 2014;60(3):196–199. doi: 10.1016/j.jcv.2014.04.002. [DOI] [PubMed] [Google Scholar]
- De Maria A., Fogli M., Costa P., Murdaca G., Puppo F., Mavilio D., et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur. J. Immunol. 2003;33(9):2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W., Duangchinda T., Lin C.L., Vasanawathana S., Jones M., Jacobs M., et al. A complex interplay among virus, dendritic cells, T cells, and cytokines in dengue virus infections. J. Immunol. 2008;181(9):5865–5874. doi: 10.4049/jimmunol.181.9.5865. [DOI] [PubMed] [Google Scholar]
- Ferlazzo G., Tsang M.L., Moretta L., Melioli G., Steinman R.M., Munz C. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J. Exp. Med. 2002;195(3):343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G., del Puerto F., Perez A.B., Sierra B., Aguirre E., Kikuchi M., et al. Association of MICA and MICB alleles with symptomatic dengue infection. Hum. Immunol. 2011;72(10):904–907. doi: 10.1016/j.humimm.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Golden-Mason L., Cox A.L., Randall J.A., Cheng L., Rosen H.R. Increased natural killer cell cytotoxicity and NKp30 expression protects against hepatitis C virus infection in high-risk individuals and inhibits replication in vitro. Hepatology. 2010;52(5):1581–1589. doi: 10.1002/hep.23896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Vaughn D.W., Kalayanarooj S., Nimmannitya S., Suntayakorn S., Nisalak A., et al. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J. Infect. Dis. 1999;179(4):755–762. doi: 10.1086/314680. [DOI] [PubMed] [Google Scholar]
- Haeberlein S., Sebald H., Bogdan C., Schleicher U. IL-18, but not IL-15, contributes to the IL-12-dependent induction of NK-cell effector functions by Leishmania infantum in vivo. Eur. J. Immunol. 2010;40(6):1708–1717. doi: 10.1002/eji.200939988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B., O'Rourke E.J. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 1977;146(1):201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B., O'Rourke E.J. Antibody-enhanced dengue virus infection in primate leukocytes. Nature. 1977;265(5596):739–741. doi: 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- Hecht M.L., Rosental B., Horlacher T., Hershkovitz O., De Paz J.L., Noti C., et al. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J. Proteome Res. 2009;8(2):712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- Hershkovitz O., Rosental B., Rosenberg L.A., Navarro-Sanchez M.E., Jivov S., Zilka A., et al. NKp44 receptor mediates interaction of the envelope glycoproteins from the West Nile and dengue viruses with NK cells. J. Immunol. 2009;183(4):2610–2621. doi: 10.4049/jimmunol.0802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hober D., Poli L., Roblin B., Gestas P., Chungue E., Granic G., et al. Serum levels of tumor necrosis factor-alpha (TNF-alpha), interleukin-6 (IL-6), and interleukin-1 beta (IL-1 beta) in dengue-infected patients. Am. J. Trop. Med. Hyg. 1993;48(3):324–331. doi: 10.4269/ajtmh.1993.48.324. [DOI] [PubMed] [Google Scholar]
- Innis B.L., Nisalak A., Nimmannitya S., Kusalerdchariya S., Chongswasdi V., Suntayakorn S., et al. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am. J. Trop. Med. Hyg. 1989;40(4):418–427. doi: 10.4269/ajtmh.1989.40.418. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Song J., Zhang Y., Li W., Zhang T., Qi S.M., et al. Short communication: longitudinal changes in peripheral blood NK cells during the first year of HIV-1 Infection in CD4Low and CD4High patient groups. AIDS Res. Hum. Retrovir. 2015;31(2):229–236. doi: 10.1089/aid.2014.0083. [DOI] [PubMed] [Google Scholar]
- Keawvichit R., Khowawisetsut L., Lertjuthaporn S., Tangnararatchakit K., Apiwattanakul N., Yoksan S., et al. Differences in activation and tissue homing markers of natural killer cell subsets during acute dengue infection. Immunology. 2018;153(4):455–465. doi: 10.1111/imm.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurane I., Innis B.L., Nimmannitya S., Nisalak A., Meager A., Ennis F.A. High levels of interferon alpha in the sera of children with dengue virus infection. Am. J. Trop. Med. Hyg. 1993;48(2):222–229. doi: 10.4269/ajtmh.1993.48.222. [DOI] [PubMed] [Google Scholar]
- Lee S., Watson M.W., Flexman J.P., Cheng W., Hammond T., Price P. Increased proportion of the CD56(bright) NK cell subset in patients chronically infected with hepatitis C virus (HCV) receiving interferon-alpha and ribavirin therapy. J. Med. Virol. 2010;82(4):568–574. doi: 10.1002/jmv.21742. [DOI] [PubMed] [Google Scholar]
- Libraty D.H., Pichyangkul S., Ajariyakhajorn C., Endy T.P., Ennis F.A. Human dendritic cells are activated by dengue virus infection: enhancement by gamma interferon and implications for disease pathogenesis. J. Virol. 2001;75(8):3501–3508. doi: 10.1128/JVI.75.8.3501-3508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.S., Yawata N., Selva K.J., Li N., Tsai C.Y., Yeong L.H., et al. The combination of type I IFN, TNF-alpha, and cell surface receptor engagement with dendritic cells enables NK cells to overcome immune evasion by dengue virus. J. Immunol. 2014;193(10):5065–5075. doi: 10.4049/jimmunol.1302240. [DOI] [PubMed] [Google Scholar]
- Luangtrakool P., Vejbaesya S., Luangtrakool K., Ngamhawornwong S., Apisawes K., Kalayanarooj S., et al. Major histocompatibility complex Class I chain-related A and B (MICA and MICB) gene, allele, and haplotype associations with dengue infections in ethnic thais. J. Infect. Dis. 2020;222(5):840–846. doi: 10.1093/infdis/jiaa134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera S., Rapp M., Firth M.A., Beilke J.N., Lanier L.L., Sun J.C. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med. 2016;213(2):225–233. doi: 10.1084/jem.20150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y., et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409(6823):1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- Matta J., Baratin M., Chiche L., Forel J.M., Cognet C., Thomas G., et al. Induction of B7-H6, a ligand for the natural killer cell-activating receptor NKp30, in inflammatory conditions. Blood. 2013;122(3):394–404. doi: 10.1182/blood-2013-01-481705. [DOI] [PubMed] [Google Scholar]
- McKechnie J.L., Beltran D., Pitti A., Saenz L., Arauz A.B., Vergara R., et al. HLA upregulation during dengue virus infection suppresses the natural killer cell response. Front. Cell Infect. Microbiol. 2019;9:268. doi: 10.3389/fcimb.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsapaya J., Dejnirattisai W., Xu X.N., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9(7):921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- Mustafa A.S., Elbishbishi E.A., Agarwal R., Chaturvedi U.C. Elevated levels of interleukin-13 and IL-18 in patients with dengue hemorrhagic fever. FEMS Immunol. Med. Microbiol. 2001;30(3):229–233. doi: 10.1111/j.1574-695X.2001.tb01575.x. [DOI] [PubMed] [Google Scholar]
- Naiyer M.M., Cassidy S.A., Magri A., Cowton V., Chen K., Mansour S., et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci. Immunol. 2017;2(15) doi: 10.1126/sciimmunol.aal5296. [DOI] [PubMed] [Google Scholar]
- Nguyen K.B., Salazar-Mather T.P., Dalod M.Y., Van Deusen J.B., Wei X.Q., Liew F.Y., et al. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 2002;169(8):4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- Pacsa A.S., Agarwal R., Elbishbishi E.A., Chaturvedi U.C., Nagar R., Mustafa A.S. Role of interleukin-12 in patients with dengue hemorrhagic fever. FEMS Immunol. Med. Microbiol. 2000;28(2):151–155. doi: 10.1111/j.1574-695X.2000.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Petitdemange C., Becquart P., Wauquier N., Beziat V., Debre P., Leroy E.M., et al. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011;7(9) doi: 10.1371/journal.ppat.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitdemange C., Wauquier N., Jacquet J.M., Theodorou I., Leroy E., Vieillard V. Association of HLA class-I and inhibitory KIR genotypes in Gabonese patients infected by Chikungunya or Dengue type-2 viruses. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders P.M., Vivian J.P., O'Connor G.M., Sullivan L.C., Pymm P., Rossjohn J., et al. A bird's eye view of NK cell receptor interactions with their MHC class I ligands. Immunol. Rev. 2015;267(1):148–166. doi: 10.1111/imr.12319. [DOI] [PubMed] [Google Scholar]
- Schepis D., Gunnarsson I., Eloranta M.L., Lampa J., Jacobson S.H., Karre K., et al. Increased proportion of CD56bright natural killer cells in active and inactive systemic lupus erythematosus. Immunology. 2009;126(1):140–146. doi: 10.1111/j.1365-2567.2008.02887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabrish S., Karnik N., Gupta V., Bhate P., Madkaikar M. Impaired NK cell activation during acute dengue virus infection: a contributing factor to disease severity. Heliyon. 2020;6(7):e04320. doi: 10.1016/j.heliyon.2020.e04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapsoba A.S.A., Djigma F.W., Bayala B., Sorgho P.A., Traore L., Zohoncon T.M., et al. KIR2DL2, KIR2DL5A and KIR2DL5B genes induce susceptibility to dengue virus infection, while KIR3DL3 and KIR2DS5 confer protection. Mediterr. J. Hematol. Infect. Dis. 2022;14(1) doi: 10.4084/MJHID.2022.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B., Burgess T.H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003;197(7):823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielens A., Vivier E., Romagne F. NK cell MHC class I specific receptors (KIR): from biology to clinical intervention. Curr. Opin. Immunol. 2012;24(2):239–245. doi: 10.1016/j.coi.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Townsley E., O'Connor G., Cosgrove C., Woda M., Co M., Thomas S.J., et al. Interaction of a dengue virus NS1-derived peptide with the inhibitory receptor KIR3DL1 on natural killer cells. Clin. Exp. Immunol. 2016;183(3):419–430. doi: 10.1111/cei.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilgenburg B., Scherwitzl I., Hutchinson E.C., Leng T., Kurioka A., Kulicke C., et al. MAIT cells are activated during human viral infections. Nat. Commun. 2016;7:11653. doi: 10.1038/ncomms11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn D.W., Green S., Kalayanarooj S., Innis B.L., Nimmannitya S., Suntayakorn S., et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 2000;181(1):2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Wagstaffe H.R., Clutterbuck E.A., Bockstal V., Stoop J.N., Luhn K., Douoguih M., et al. Ebola virus glycoprotein stimulates IL-18-dependent natural killer cell responses. J. Clin. Investig. 2020;130(7):3936–3946. doi: 10.1172/JCI132438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 1997. Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control.https://iris.who.int/handle/10665/41988 Available from. [Google Scholar]
- Yenchitsomanus P.T., Sricharoen P., Jaruthasana I., Pattanakitsakul S.N., Nitayaphan S., Mongkolsapaya J., et al. Rapid detection and identification of dengue viruses by polymerase chain reaction (PCR) Southeast Asian J. Trop. Med. Public Health. 1996;27(2):228–236. [PubMed] [Google Scholar]
- Zimmer C.L., Cornillet M., Sola-Riera C., Cheung K.W., Ivarsson M.A., Lim M.Q., et al. NK cells are activated and primed for skin-homing during acute dengue virus infection in humans. Nat. Commun. 2019;10(1):3897. doi: 10.1038/s41467-019-11878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.