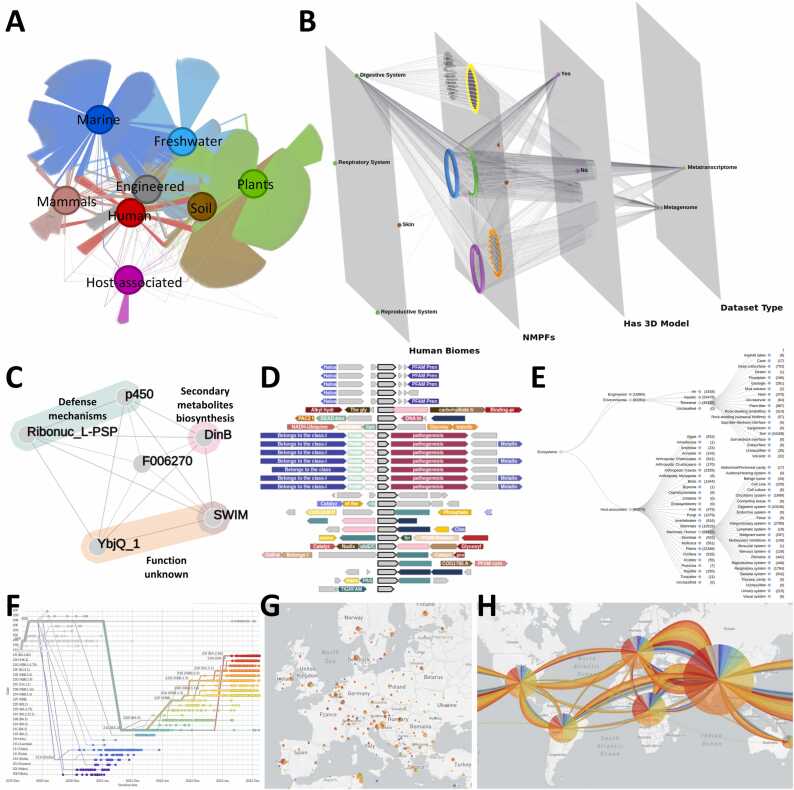
Fig. 5.
(A-C) Various network visualization schemes for data retrieved from NMPFamsDB. (A) 2D Network visualization of NMPF distribution across different biomes, rendered using Gephi. (B) 3D, multi-layered network visualization of NMPFs associated with 4 human microbiomes, as well as additional annotation (sample type and availability of 3D model), created using Arena3Dweb. (C) A gene co-occurrence network describing the gene neighborhood of a novel metagenome protein family (F006270), constructed with data from NMPFamsDB and rendered using NORMA. The functional annotation of F006270’s neighboring genes is presented in the form of colored groups. (D) Gene neighborhood visualization for multiple MAGs through synteny conservation analysis, rendered using GeCoViz and the FESNov catalog. (E) Tree visualization of metagenome ecosystems, using the GOLD classification system. The number of metagenomic datasets associated with each ecosystem is given in parentheses. (F) Chronological progression of different SARS-Cov-2 strains in the form of a histogram, rendered using NextStrain. (G-H) Map visualizations of the geographical distribution across Europe (G) and global dispersion patterns of COVID-19 (H) rendered using NextStrain.