Figure 1. Acb2 from phage PaMx33 binds cyclic trinucleotides and 3’, 2’-cGAMP.
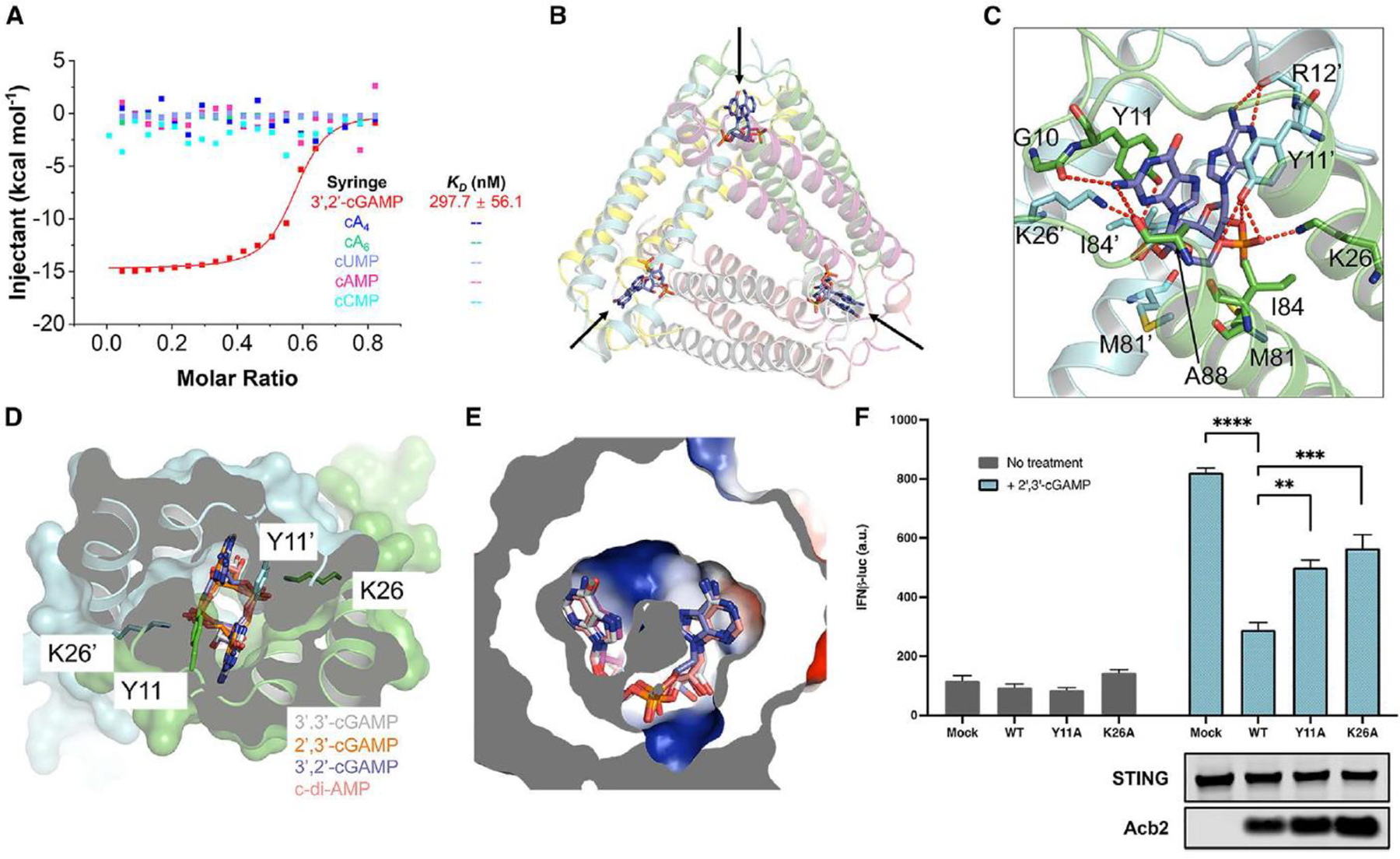
(A) ITC assays to test binding of cyclic nucleotides to PaMx33-Acb2. Representative binding curves and binding affinities are shown. The KD values are mean ± s.d. (n=3). Raw data for these curves are shown in Figure S2.
(B) Overall structure of Acb2 complexed with 3’,2’-cGAMP, which are indicated by arrows.
(C) Detailed binding between Acb2 and 3’,2’-cGAMP. Residues involved in 3’,2’-cGAMP binding are shown as sticks. Red dashed lines represent polar interactions.
(D) Structural alignment among 3’,2’-cGAMP, 2’,3’-cGAMP, 3’,3’-cGAMP and c-di-AMP bound Acb2. Surface representation overlaid to cartoon representation, highlighting the binding pocket of CDNs.
(E) Electrostatic surface model showing the binding pocket of CDNs. The CDNs are colored as in D.
(F) 293T-Dual cells were transfected with hSTING and Acb2 or its mutants, then treated with 2’,3’-cGAMP. STING activation was read as luciferase signal controlled by an interferon promoter. A western blot is shown probing the expression of STING and Acb2.
