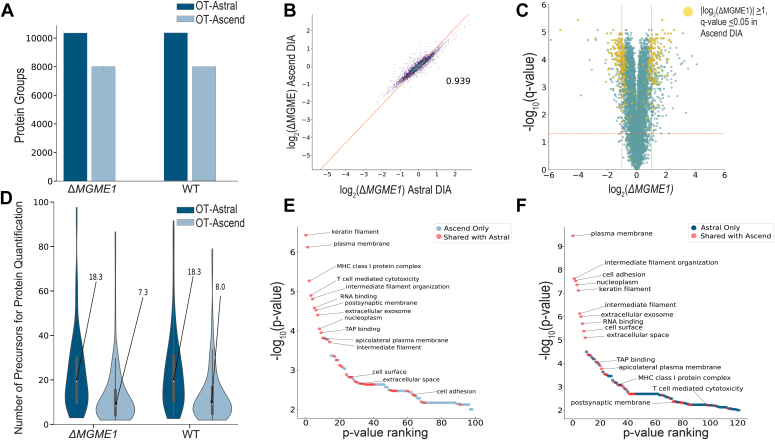
Fig. 4.
Comparison of differential expression analyses upon CRISPR-Cas9 MGME1 KO between 30-min Orbitrap Astral and 61-min Orbitrap Ascend DIA methods.A, unique WT and ΔMGME1 KO HAP1 protein groups identified across methods. B, correlation of log2-transformed fold changes measured by OT-Ascend and that measured by OT-Astral have a Pearson correlation coefficient of 0.939 (n = 7304). C, two-sided t tests for each quantified protein present in three KO and three WT replicates (n = 9462) were performed assuming equal variance. Negative log10-transformed q-values were plotted against log2-transformed fold changes between KO and WT for the OT-Astral-quantified proteins (green and yellow). OT-Astral delivered 449 significantly upregulated or downregulated proteins. Subsequently, 234 significantly changed proteins from the OT-Astral dataset were also determined to be significantly upregulated or downregulated by the OT-Ascend method (yellow). D, subset of proteins that pass the significance threshold within OT-Ascend dataset but not the OT-Astral dataset (143 of 380) have a higher median number of precursors that were used for protein quantitation in the Astral dataset. E-F, GO-term enrichment was performed on proteins quantified in all three KO and WT replicates that were determined to have a fold-change of at least two and a q-value <0.05. There were 40 shared significantly enriched (p-value <0.01) GO-terms (red), and the top 11 enriched in each dataset were corroborated in the comparison dataset. DIA, data-independent acquisition; GO, gene ontology.