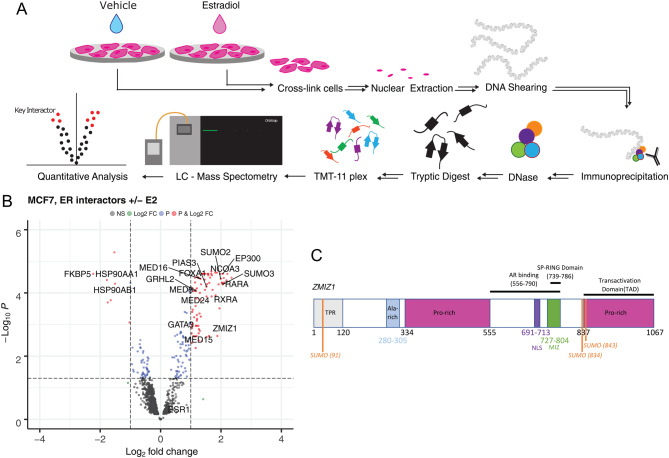
Figure 1.
qPLEX-RIME of ER activation identifies novel co-factor ZMIZ1. (A) qPLEX-RIME enables the quantitative comparison of multiple conditions to identify key interactions between transcription factors on the chromatin. We stimulated ER-positive breast cancer cell lines with estradiol and undertook a comparative analysis against an unstimulated control to identified the changes in the ER interactome on activation. (B) ER interacting proteins identified by qPLEX-RIME in MCF7. Top ranking differential-associated proteins detected with P< 0.05 and LogFC >1 are highlighted in red. Gain of known co-factors GATA3 (Theodorou et al. 2013), RARA (Ross-Innes et al. 2010), EP300, GRHL2 (Holding et al. 2019) and NCOA3 were all detected along with the loss of HSP90 on binding of estradiol by the ER. Parts of the mediator complex (MED8, MED16 and MED24), along with pioneer factor FOXA1 were also detected (Supplementary Table 1). ZMIZ1 has not previously been reported to interact with the ER. (C) A schematic presentation of the ZMIZ1 protein. The AR binding region was previously identified by the interaction of a 556-790aa truncated mutant with an AR-GAL4-DBD fusion protein leading to activation of β-gal reporter gene. The C-terminal, proline-rich region of ZMIZ1, was identified as an intrinsic transactivation domain (TAD) (Sharma et al. 2003). TPR, tetratricopeptide repeat (Wang et al. 2018); NLS, nuclear localisation signal (Sharma et al. 2003). A full colour version of this figure is available at https://doi.org/10.1530/JME-23-0133.

 This work is licensed under a
This work is licensed under a