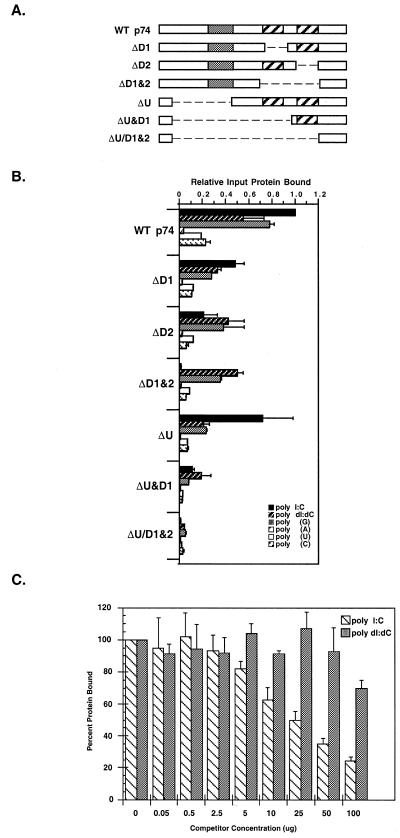
Figure 5.
Nucleotide binding analysis of the p74 protein. (A) Deletion constructs. The domain structure of wild-type p74 and various deletion mutants are shown with the USCR (gray) and dsRBMs (hatched) as noted. (B) Polynucleotide binding analysis. p74 was in vitro translated in rabbit reticulocyte lysates and the 35S-labeled translation products were incubated with various excess polynucleotide resins (as noted). Bound proteins were eluted and the amount of bound protein was determined relative to wild-type binding (see Materials and Methods). (C) Competition binding studies. In vitro translated wild-type p74 was incubated with various concentrations of competitor nucleic acid [either poly(I:C) or poly(dI:dC), as indicated] followed by incubation with dsRNA–agarose. Bound proteins were eluted and analyzed as in (B).