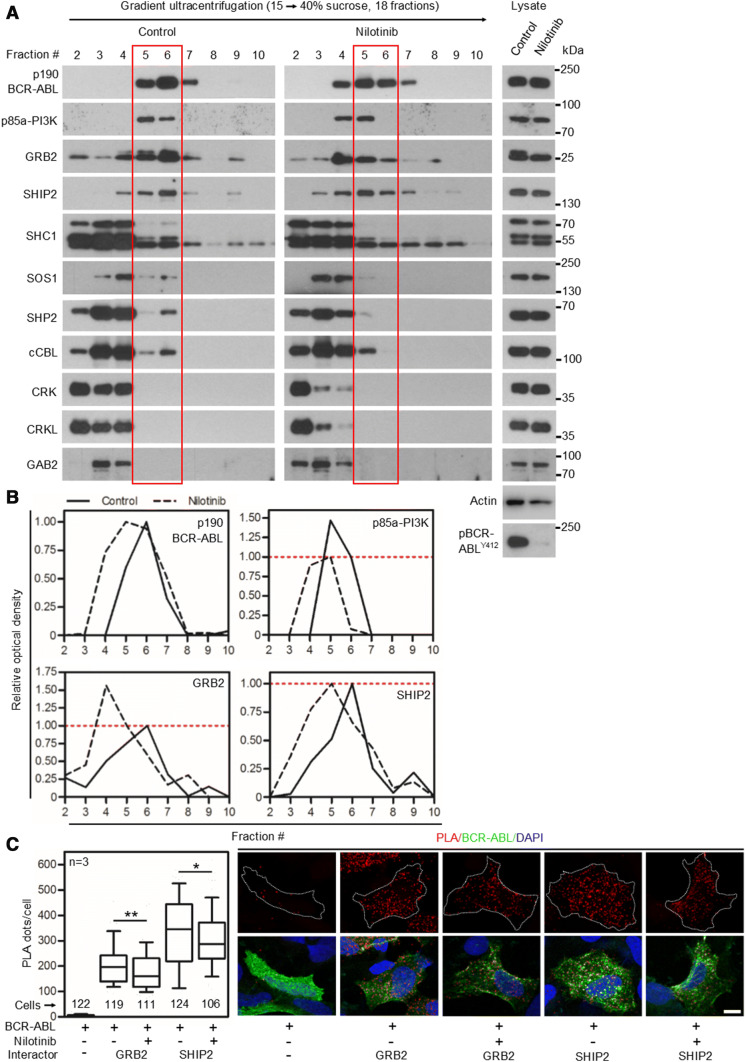
Fig. 1.
Nilotinib causes partial dissolution of BCR–ABL signaling complex. a 293T cells were transfected with p190 BCR–ABL, native cell lysates were subjected to ultracentrifugation in the 15–40% sucrose gradient, and collected fractions were analyzed by western blot. The presence of BCR–ABL signal in more than one fraction suggests the existence of complexes of different compositions. Note the various degrees of co-sedimentation of BCR–ABL with p85a-PI3K, GRB2, SHIP2, SHC1, SOS1, SHP2 and cCBL; no co-sedimentation with CRK, CRKL or GAB2 was found. Inhibition of BCR–ABL kinase activity with 100 nM nilotinib resulted in a shift of a fraction of the BCR–ABL complexes towards lighter fractions, suggesting partial dissolution of the BCR–ABL signaling complex. b The western blot analysis of proteins co-sedimenting with BCR–ABL (p85a-PI3K, GRB2 and SHIP2) was quantified as described in “Materials and methods”. Note that portion of GRB2, but not SHIP2 or p85a-PI3K dissociated from the BCR–ABL complex after nilotinib treatment. Data represent a single experiment out of three independent experiments carried out. The fractions containing most of the p190 BCR–ABL are highlighted in red. Phosphorylation (p) at ABL Y412 was used to determine the degree of BCR–ABL inhibition using nilotinib; actin serves as a loading control in total cell lysates used for ultracentrifugation. c Cells were transfected with FLAG-tagged p190 BCR–ABL, V5-tagged GRB2 or SHIP2, treated with nilotinib, and subjected to PLA. The antibodies against protein tags were used in PLA (red); cABL antibody was used to counterstain the transfected cells (green). Cells transfected with BCR–ABL and an empty vector serve as the negative control. Number of PLA dots per cell was calculated and graphed (10–90 percentile). Statistically significant differences were highlighted (Student’s t test with Welch’s correction for unequal variances; *p < 0.05, **p < 0.01). Scale bars, 10 µm