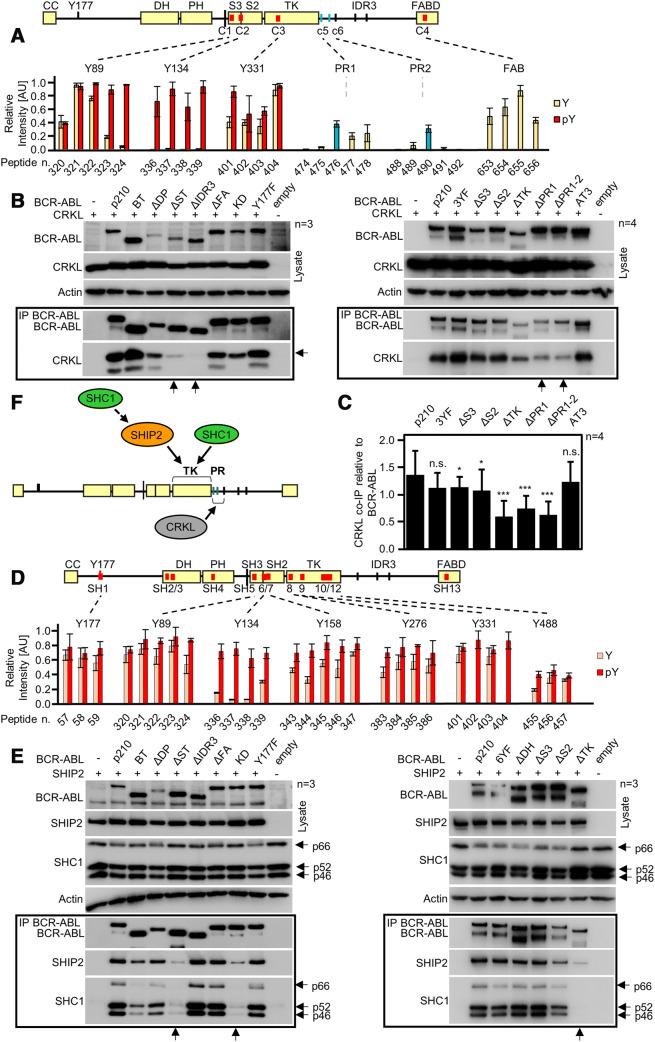
Fig. 6.
Interaction of CRKL and SHIP2 with BCR–ABL. a BCR–ABL scheme with red lines indicating potential CRKL binding sites identified by microarray. Graph shows binding intensities for phosphorylated (red) and non-phosphorylated peptides; strong binding is shown for phosphorylated Y89, Y134 and Y331. Sites c5 (PR1) and c6 (PR2) are not formally considered; however, blue bars indicate positive binding for respective peptides. b Co-immunoprecipitation (co-IP) of BCR–ABL with CRKL in 293T cells. Deletion of IDR3 and both PR1 and PR2 sites (constructs ΔPR1 and ΔPR1-2) limits the interaction of CRKL with BCR–ABL (arrows). Substituting Y89, Y134 and Y331 to phenylalanines (3YF) produced no effect on CRKL binding. c Quantification of CRKL co-IP with BCR–ABL constructs from (b, right panel). CRKL was normalized to BCR–ABL levels, error bars indicate SD from four independent experiments (n = 4). Statistically significant differences are indicated (Student’s t test, *p < 0.05, ***p < 0.001; n.s., non-significant). d Microarray analysis of SHIP2 binding to BCR–ABL shows multiple binding sites in the SH3–SH2–TK domains, association with the tyrosine-phosphorylated motifs is indicated in red. Binding site for Y488 was obtained in independent experiment and, therefore, different threshold applied to this site. e Immunoprecipitation of BCR–ABL constructs with SHIP2 in transfected 293T cells. Deletion of SH3–SH2–TK (ΔST), or TK domain (ΔTK) limits SHIP2 binding (arrows). Substituting Y89, Y134, Y158, Y276, Y331 and Y488 to phenylalanines (6YF) has no effect on SHIP2 binding. SHC1 associated with BCR–ABL in a manner similar to SHIP2, suggesting mutual interaction. f Scheme of proposed interaction. CRKL binds to region containing PR1 and PR2. SHIP2 binds to the TK region of ABL and also recruits SHC1 to BCR–ABL. n number of independent experiments