Fig. 8.
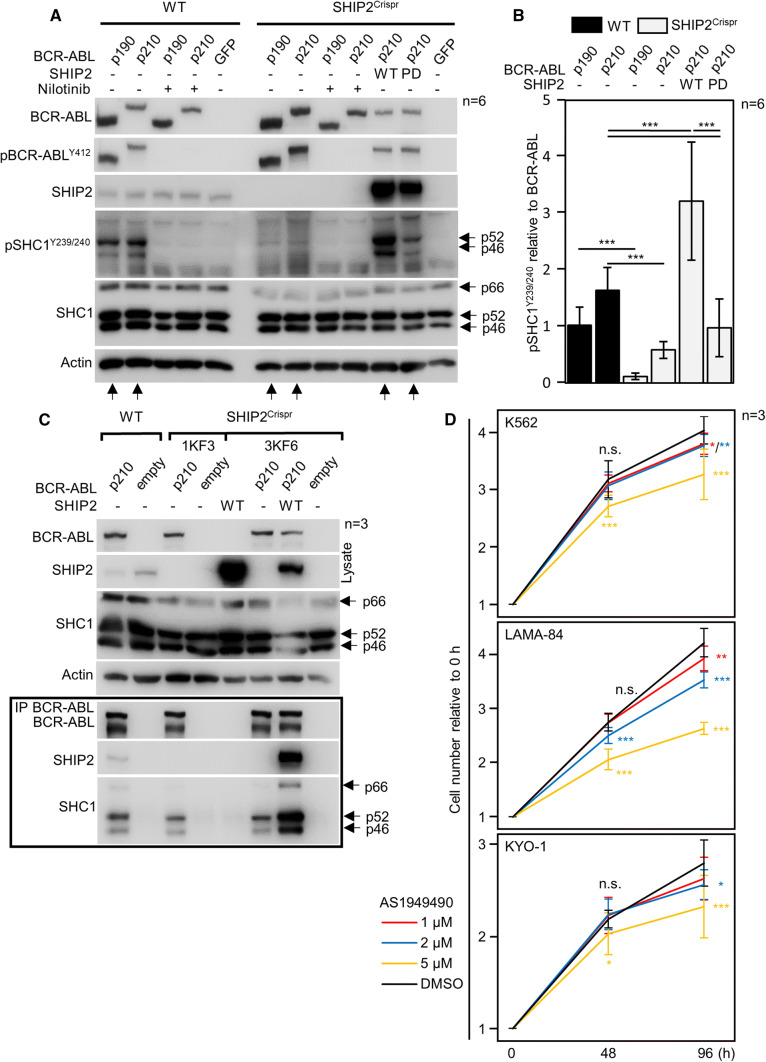
SHIP2 is required for BCR–ABL-mediated phosphorylation of SHC1. a, b Wild-type (wt) 293T cells and cells with SHIP2 deleted by CRISPR/Cas9 (SHIP2Crispr) were transfected by BCR–ABL, and analyzed for presence and phosphorylation (p) of given proteins by western blot. In contrast to wt cells, BCR–ABL could not phosphorylate SHC1 in SHIP2Crispr cells. Addition of wt SHIP2 into the SHIP2Crispr cells fully rescued BCR–ABL-mediated SHC1 phosphorylation, while the addition of catalytically inactive (phosphatase-dead, PD) had only minor effect. Same results were obtained with two independent SHIP2Crispr cell lines (not shown). b Quantification of SHC1 phosphorylation in (a), normalized to BCR–ABL levels. Bars represent averages from six experiments (n) with indicated SD, statistically significant differences are indicated (Student’s t test, ***p < 0.001). c SHC1 interacts with BCR–ABL in SHIP2Crispr 293T cells. BCR–ABL was transfected to wt and two clones of SHIP2Crispr 293T cells (1KF3, 3KF6), and immunoprecipitated (IP). SHC1 co-immunoprecipitates with BCR–ABL in SHIP2Crispr, suggesting that lack of SHC1 phosphorylation in SHIP2Crispr cells is not due to loss of interaction with BCR–ABL. Data are representative for 3 independent experiments (n). d K562, LAMA-84 and KYO-1 cells were seeded at 500 cells/μl, and treated with AS1949490 or DMSO. Cells were counted 48 and 96 h after treatment. Data represent averages ± SD from three independent experiments (n), with two technical replicates for each treatment. Statistically significant differences are indicated (Student’s t test, *p < 0.05, ***p < 0.001)