Fig. 1.
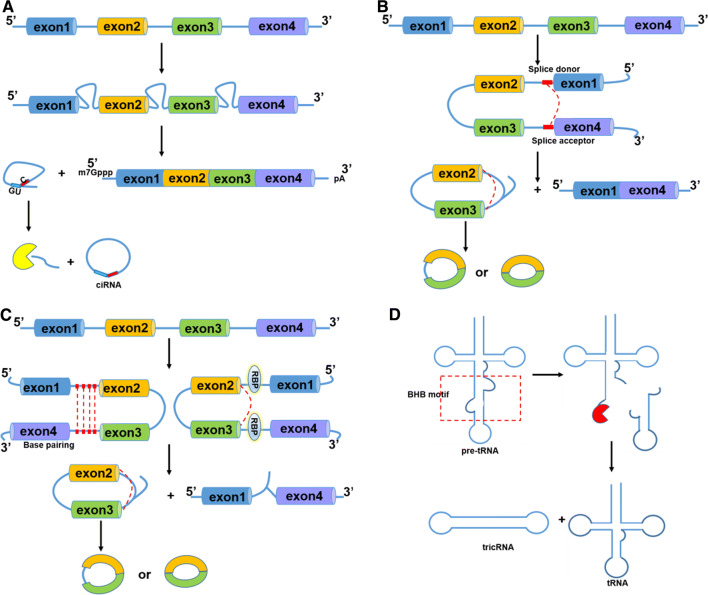
Biogenesis of circRNA. a Canonical splicing. Pre-mRNA is spliced by spliceosome to remove intervening introns and leave only exons sequentially connected to form mature mRNA. The rest lariat intron can escape debranching and degradation with 7 nt GU-rich sequences near the 5′ splice site and 11 nt C-rich sequence near branch site and instead the 3′ “tail” downstream sequence is trimmed to form stable ciRNA. b Lariat-driven circularization. A downstream 5′ splice site of exon 4 is joined to an upstream 3′ splice site of exon 1 forming lariat structure. And then, the lariat containing skipped exons undergoes internal splicing of one intron to form EIciRNA or splicing of two introns to form ecircRNA. c Intron-pairing-driven circularization. Intronic motifs flanking circularized exon 2 and 3 have considerably complementary sequences which bring splicing sites close to form EIciRNA or eciRNA. RNA binding proteins (RBPs) driven circularization. RBPs with binding sites on flanking introns of exons 2 and 3 could bring two splicing sites close together to facilitate circularization. d The tRNA splicing endonuclease complex (TSEN) recognizes bulge–helix–bulge (BHB) motif and carries out intron excision of pre-tRNA and then release and ligate the end to form tRNA and tricRNA