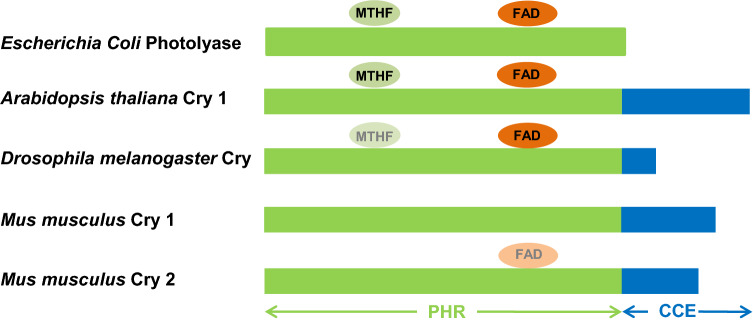
Fig. 3.
Schematic representation of the structures of photolyase of bacteria (Escherichia coli) and representative cryptochromes (CRY) of plant (Arabidopsis thaliana), insects (type I CRY of Drosophila melanogaster), and mammals (type II CRY of mice, Mus musculus), with approximate binding sites of 5-methyltetrahydrofolate (MTHF) (equivocal in Drosophila) and flavin adenine dinucleotide (FAD) cofactors (equivocal in mammalian CRY 2). PHR photolyase homology region domain, CCE C-terminal extension domain [23–25]