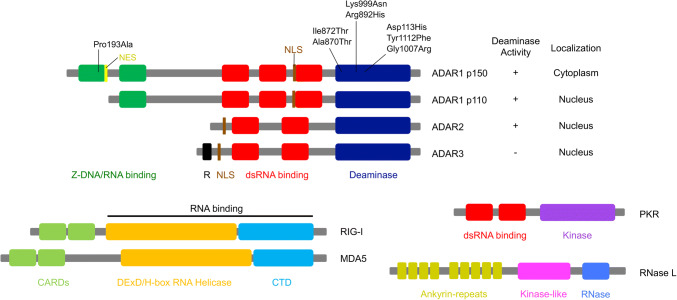
Fig. 1.
Structural representation of ADARs and RNA-binding proteins involved in dsRNA-sensing pathways. Cytoplasmic adenosine deaminase acting on RNA 1 (ADAR1) p150 comprises two Z-DNA/RNA binding domains (green), three double-stranded (ds)RNA-binding domains (red), and a deaminase domain (dark blue), while nuclear ADAR1 p110 is a truncated isoform that lacks a Z-DNA/RNA-binding domain. A nuclear localization signal (NLS; shown in brown) is present in both p150 and p110 isoforms, whereas a nuclear export signal (NES; shown in yellow) is present in the p150 isoform only. Both ADAR2 and ADAR3 are composed of two dsRNA-binding domains and a deaminase domain, and are located in the nucleus. ADAR3, which contains arginine-rich domain (R; shown in black), has not been shown to have editing activity. Amino acid substitutions resulting from point mutations in the ADAR1 gene, identified in patients with Aicardi–Goutières syndrome (AGS), are also shown. Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) are members of RIG-I-like receptors and comprise two caspase activation and recruitment domains (CARDs; shown in light green), which mediate signal transduction through interaction with the mitochondrial anti-viral-signaling protein (MAVS) with a DExD/H-box RNA helicase domain (orange) and a C-terminal domain (CTD; shown in light blue), both of which are required for RNA binding. Protein kinase R (PKR) is composed of two dsRNA-binding domains (red) and a kinase domain (purple). RNase L comprises nine ankyrin-repeats domain (dark yellow), a kinase-like domain (pink) and an RNase domain (blue). An ankyrin-repeats domain contains the site for binding to 2′,5′-oligoadenylates, which is produced by oligoadenylate synthetase (OAS) proteins