Abstract
Cytidine base editors (CBEs) have been demonstrated to be useful for precisely inducing C:G-to-T:A base mutations in various organisms. In this study, we showed that the BE4-Gam system induced the targeted C-to-T base conversion in porcine blastocysts at an efficiency of 66.7–71.4% via the injection of a single sgRNA targeting a xeno-antigen-related gene and BE4-Gam mRNA. Furthermore, the efficiency of simultaneous three gene base conversion via the injection of three targeting sgRNAs and BE4-Gam mRNA into porcine parthenogenetic embryos was 18.1%. We also obtained beta-1,4-N-acetyl-galactosaminyl transferase 2, alpha-1,3-galactosyltransferase, and cytidine monophosphate-N-acetylneuraminic acid hydroxylase deficient pig by somatic cell nuclear transfer, which exhibited significantly decreased activity. In addition, a new CBE version (termed AncBE4max) was used to edit genes in blastocysts and porcine fibroblasts (PFFs) for the first time. While this new version demonstrated a three genes base-editing rate of 71.4% at the porcine GGTA1, B4galNT2, and CMAH loci, it increased the frequency of bystander edits, which ranged from 17.8 to 71.4%. In this study, we efficiently and precisely mutated bases in porcine blastocysts and PFFs using CBEs and successfully generated C-to-T and C-to-G mutations in pigs. These results suggest that CBEs provide a more simple and efficient method for improving economic traits, reducing the breeding cycle, and increasing disease tolerance in pigs, thus aiding in the development of human disease models.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03205-2) contains supplementary material, which is available to authorized users.
Keywords: Base editing, Cytidine base editors (CBEs), Pigs, BE4-Gam, AncBE4max
Introduction
In recent years, the CRISPR–Cas9 system has been widely used as a method for targeted genome editing in many animals, plants, and prokaryotes, including pigs [1–3], rabbits [4–6], dogs [7], mice [8, 9], rice [10, 11], and bacteria [12], and thus serves as a launchpad for a new era in site-specific genome editing. Currently, the CRISPR–Cas9 system serves as a powerful tool for generating human disease models by knocking out disease-related genes. With the development of DNA sequencing, the vast majority of human genetic diseases are induced by point mutations, rather than insertions or deletions (indels) [13–16]. Hence, genome editing with programmable nucleases, such as Cas9, to mutate a single base is technologically challenging due to the high frequency of random indels and low efficiency of base mutations [16, 17]. More recently, an effective base editor that combines cytidine deaminase or adenine deaminase with the CRISPR/Cas9 platform has been developed for precise C-to-T or A-to-G conversions without requiring a template-donor plasmid or double-stranded breaks (DSBs) in targeted genomic DNA. These editors have been broadly used to precisely edit target bases in human embryos [18] rabbits [4], pigs [19] mice [20], zebrafish [21], and many plants [10, 11, 22, 23], and have enormous potential in therapeutic applications [24–26].
Significant improvements in the quality of human life have also substantially improved the human life expectancy, resulting in increased risks of chronic diseases and end-stage organ failure [27] and acute organ resource shortages.
The use of domestic pigs as livestock has been widely studied over the past few years. Many genetically modified pigs have been generated to improve their economic traits, such as production yield [28–31], growth efficiency [32], and disease resistance [33–37]. Furthermore, pigs that share similar genetic, physiological, and anatomical features as well as a similar body size with humans are regarded as important candidate organ donors for xeno-transplantation and as an important material for human disease models [38–40].
At present, the major hyperacute rejection (HAR)-related xeno-antigen [41] comprises alpha-1,3-galactose (alpha-gal), the cell surface glycan antigen Sd(a) and the cell surface molecule sialic acid, Neu5Gc (N-glycolylneuraminic acid), whose syntheses are related to alpha-1,3-galactosyltransferase (encoded by the GGTA1 gene), beta-1,4-N-acetyl-galactosaminyl transferase 2 (encoded by the B4GalNT2 gene), and cytidine monophosphate-N-acetylneuraminic acid hydroxylase (encoded by the CMAH gene), respectively. Although there is a report of GGTA1/B4GalNT2/CMAH three gene knock out in pigs generated by Cas9 nuclease-induced DNA DSB [41], a recent study shows that it may produce excess undesired editing by-product included unexpected chromosomal truncated and cause unwanted dysfunctional protein [41–44]. The pigs produced by Cas9 nuclease-induced DNA DSB may have potential risks for xeno-transplantation. Therefore, the preparation of multiple xeno-antigen-related gene-deficient pigs with lower risk is urgently needed.
To our knowledge, this is the first to knock out a xenogeneic antigen-related gene in the porcine genome using CBEs by introducing nonsense mutations, which suggests the enormous potential of using CBEs to improve economic traits, reduce the breeding cycle time, increase disease tolerance, and develop human disease models in pigs. Furthermore, we demonstrated that the new CBE version (AncBE4max) functions as a porcine genome base editor and exhibits a base-editing efficiency higher than those of BE4-Gam in porcine blastocysts and porcine fibroblasts (PFFs). Overall, we illustrated that CBEs are a simple and efficient method for introducing targeted base edits in porcine embryos and fibroblasts and laid the foundation for producing human disease models and provide a possible treatment for genetic diseases.
Results
Base mutation of GGTA1, B4galNT2, and CMAH in porcine blastocysts and porcine fibroblasts with BE4-Gam systems
To determine the efficiency of CBEs in the porcine genome, we attempt to accomplish three xeno-antigens corresponding gene-deficient with CBEs in porcine blastocysts and fibroblasts.
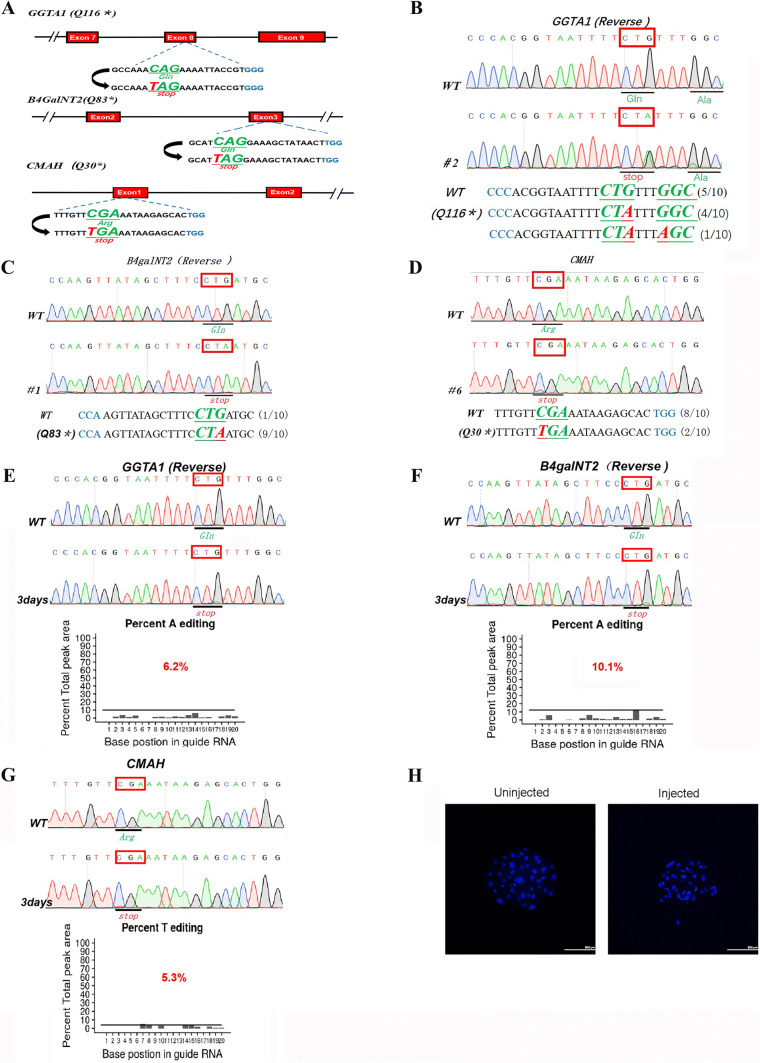
The catalytic domain of alpha-1,3-galactosyltransferase is located in exon 8, and exon 9 of GGTA1 and human cytidine monophosphate-N-acetylneuraminic acid hydroxylase is inactivated due to the loss of 104 amino acids within the N-terminus of cytidine monophosphate-N-acetylneuraminic acid hydroxylase. Therefore, our target sites for exploring whether CBEs can precisely edit bases at target loci in the porcine embryo and fibroblast genomes were based on exon 8 (Q116*) of GGTA1, exon 1 (Q30*) of CMAH and exon 3 (Q83*) of B4galNT2 (Fig. 1a). We injected BE4-Gam mRNA and targeting single-guide (sgRNAs) into porcine parthenogenetic embryos and performed Sanger sequencing and T-A cloning at 7 days later, revealing that CBEs successfully introduced base mutations at each of the target sites (Fig. 1b–d; Supplementary Figure 1). The mutation rates ranged from 66.7 to 71.4%, and the efficiency of bystander editing ranged from 11.1 to 33.3%; no indels or C to non-T mutations were induced (Table 1).
Fig. 1.
Single-gene base mutation of GGTA1, B4galNT2, CMAH in porcine blastocysts, and PFFs with BE4-Gam system. a The target sequences at the GGTA1, B4galNT2, and CMAH locus. The PAM sequence, sgRNA-target sequences, and substituted bases are shown in blue, black, and red, respectively. Representative sequencing chromatograms at the GGTA1 (b), B4galNT2 (c), and CMAH (d) targets of WT and edited porcine blastocysts. Target sequence (black), PAM region (blue), target sites (red), and mutant amino acid (bold and italic). Representative sequencing chromatograms at the GGTA1 (e), B4galNT2 (f), and CMAH (g) targets of WT and edited PFFs. Target amino acid are indicated by red box. The efficiency of mutation was predicted with online tool EditR (https://moriaritylab.shinyapps.io/editr-master). h The blastocysts of un-injected blastocysts and the blastocysts which injected single sgRNAs plus BE4-Gam. The blastocysts was stained with DAPI, and the nucleus in the blastocyst was shown in blue dots
Table 1.
Summary of base-editing rates using BE4-Gam system in porcine embryos by injecting single sgRNA
| Target genes | No. of zygotes | No. of 2-cell (%) | No. of blastocysts (%) | No. of mutants (%) | No. of target mutants (%) | No. of bystander editing (%) | No. of indels (%) | No. of C to non-T (%) |
|---|---|---|---|---|---|---|---|---|
| GGTA1 | 60 | 17 (28.3) | 6 (10.0) | 4/6 (66.7) | 4/6 (66.7) | 2/6 (33.3) | 0/6 (0) | 0/6 (0) |
| B4GalNT2 | 60 | 22 (36.7) | 9 (15.0) | 6/9 (66.7) | 6/9 (66.7) | 1/9 (11.1) | 0/9 (0) | 0/9 (0) |
| CMAH | 60 | 19 (31.7) | 7 (11.7) | 5/7 (71.4) | 5/7 (71.4) | 0/7 (0) | 0/7 (0) | 0/7 (0) |
In addition, we test whether the target base mutation incorporates successfully in each target site of Bama miniature pig fibroblasts genomes. By transfecting target sgRNA and BE4-Gam expression plasmid into Bama miniature pig fibroblasts, genome DNA was extracted at 3 days later. Results from Sanger sequencing demonstrated that it can also introduce a precise base mutation in each target site and the target site mutation rate ranges from 5.3 to 10.1% (Fig. 1e–g). Furthermore, the blastocyst development was not affected between the injected blastocysts and the un-injected blastocysts (Fig. 1h). These results indicated that this base-editing system can efficiently introduce single-gene base conversion in porcine blastocysts and PFFs.
Introducing C-to-T mutations in multiple porcine genes using the BE4-Gam system
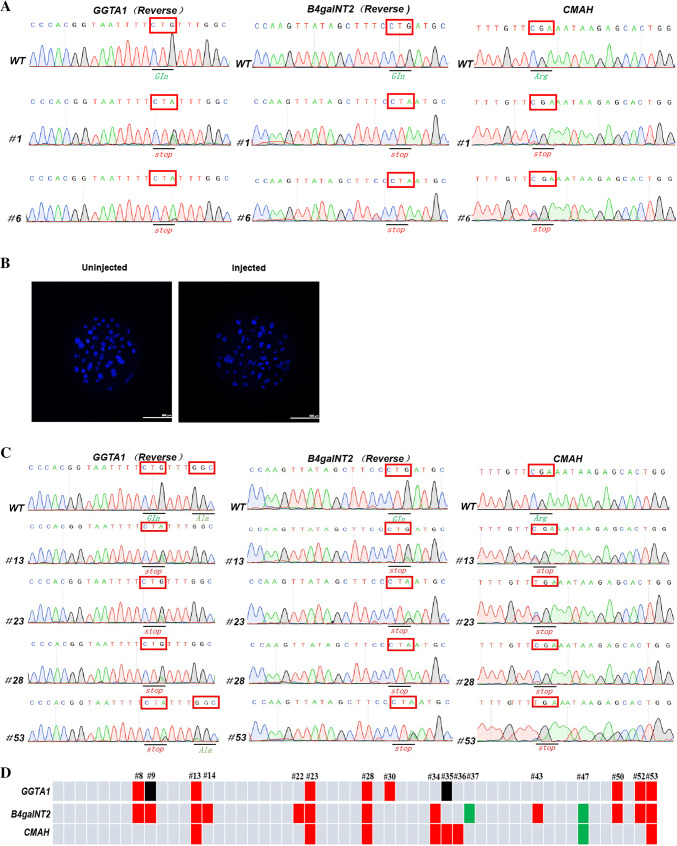
To date, most human genetic disease models have been generated by disrupting the expression of a single gene. However, in many cases, disease is determined by interactions among multiple genes [45–49]. To evaluate the ability of this base editor to mutate bases in multiple porcine genes, we simultaneously injected sgRNAs targeting three genes, GGTA1, B4galNT2, and CMAH, and BE4-Gam mRNA into porcine parthenogenetic embryos, and then evaluated the gene mutation rates. Seven days after injection, we lysed the well-developed blastocysts to obtain a DNA template for genotyping, and the Sanger sequencing and T–A cloning results showed that the BE4-Gam simultaneously mutated the three target genes (Fig. 2a). The efficiency of base editing ranged from 36.4 to 45.5% (Table 2), and target point mutations in the GGTA1, B4galNT2, and CMAH genes were observed in 5 out of 11 (45.5%) blastocysts, 5 out of 11 (45.5%) blastocysts, and 4 out of 11 (36.4%) blastocysts, respectively. Importantly, 2 out of 11 (18.1%) blastocysts exhibited three gene mutations at the target sites and 3 out of 11 (27.3%) blastocysts exhibited two gene mutations at the target sites.
Fig. 2.
Three gene C-to-T mutations in porcine blastocysts and fibroblasts with BE4-Gam system. a Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and three gene-edited porcine blastocysts. Target amino acids are indicated by red box. b The blastocysts of un-injected blastocysts and the blastocysts which injected three sgRNAs plus BE4-Gam simultaneously. The blastocysts were stained with DAPI, and the nucleus in the blastocyst was shown in blue dots. c Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and three gene-edited PFFs after transfected these three sgRNAs and BE4-Gam expression plasmid into Bama miniature PFFs. Target amino acid are indicated by red box. d Schematic of obtained PFF cell colonies. The colonies harbored C-to-T, C-to-G, and C-to-A mutations in each target gene are highlighted in red, black, and green, respectively
Table 2.
Summary of base-editing rates using BE4-Gam system in porcine embryos by injecting three sgRNAs
| Target genes | No. of zygotes | No. of 2-cell (%) | No. of blastocysts (%) | No. of mutants (%) | No. of target mutants (%) | No. of bystander editing (%) | No. of indels (%) | No. of C to non-T (%) | No. of double-gene mutants (%) | No. of three gene mutants (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| GGTA1 | 95 | 29 (30.5) | 11 (11.4) | 5/11 (45.5) | 5/11 (45.5) | 2/11 (18.1) | 0/11 (0) | 1/11 (9.9) | 3/11 (27.3) | 2/11 (18.1) |
| B4GalNT2 | 6/11 (54.5) | 5/11 (45.5) | 1/11 (9.9) | 0/11 (0) | 2/11 (18.1) | |||||
| CMAH | 4/11 (36.4) | 4/11 (36.4) | 0/11 (0) | 0/11 (0) | 2/11 (18.1) |
Although some reports have shown that the BE4-Gam system, which fuses the bacteriophage Mu-originated Gam protein and two uracil-DNA glycosylase inhibitor (UGI) to the N- and C-termini of BE3, respectively, can decrease the ratio of unwanted mutations, we observed unintended point mutations (C to non-T mutations) after injection of the three sgRNAs into porcine parthenogenetic embryos (Supplementary Figure 2; Table 2). Nevertheless, no indel events occurred in the porcine blastocyst genome. Similarly, blastocyst development was normal between the injected and un-injected blastocysts (Fig. 2b).
At the same time, we transfected these three sgRNAs and a BE4-Gam expression plasmid into Bama miniature pig fibroblasts. After 3 days, the electroporated PFFs were plated via limiting dilution, and single-cell colonies were picked 9 days later. A total of 53 colonies were obtained, and the overall mutation rates were determined by Sanger sequencing (Fig. 2c; Table 3). In total, 14 out of 53 (26.4%), 10 out of 53 (18.8%), and 8 out of 53 (15.1%) colonies had mutations within the B4galNT2, GGTA1, and CMAH genes, respectively. Based on these results, 4 out of 53 colonies contained C-to-T mutations in all three genes simultaneously (Fig. 2d). These results demonstrated that BE4-Gam can also introduce targeted mutations at multiple sites in porcine embryos and PFFs.
Table 3.
Summary of base-editing rates using BE4-Gam system in PFFs by transfecting three sgRNAs simultaneously
| Target genes | No. of mutants (%) | No. of target mutants (%) | No. of bystander editing (%) | No. of indels (%) | No. of C to non-T (%) | No. of double genes mutants (%) | No. of three genes mutants (%) |
|---|---|---|---|---|---|---|---|
| GGTA1 | 10/53 (18.8) | 10/53 (18.8) | 2/53 (3.77) | 0/53 (0) | 2/53 (3.77) | 7/53 (13.2) | 4/53 (7.5) |
| B4GalNT2 | 14/53 (26.4) | 14/53 (26.4) | 0/53 (0) | 0/53 (0) | 2/53 (3.77) | ||
| CMAH | 8/53 (15.1) | 8/53 (15.1) | 0/53 (0) | 0/53 (0) | 1/53 (1.88) |
Generating GGTA1 and B4galNT2 gene mutations in pigs by injecting three target sgRNAs and BE4-Gam mRNA into porcine IVF embryos
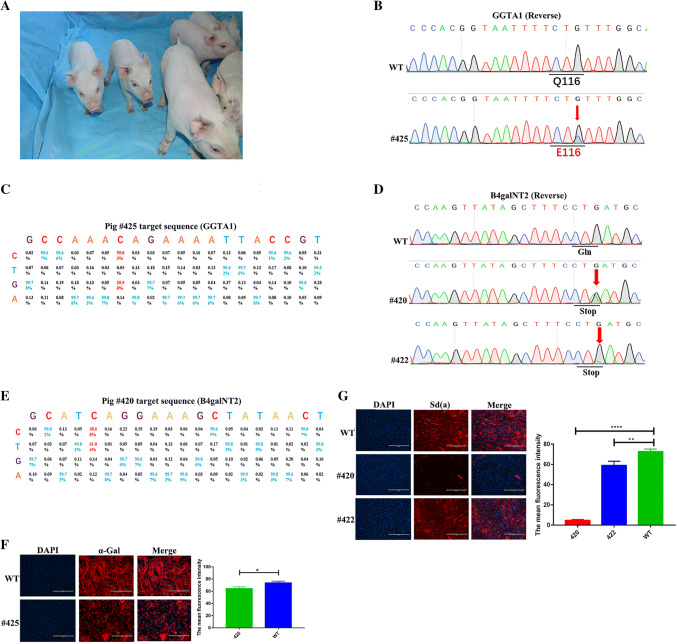
We next aimed to obtain xeno-antigen pigs deficient in these three genes for use as xeno-transplantation donors. After injecting three target sgRNAs and BE4-Gam mRNA into 700 in vitro fertilization (IVF) embryos, the embryos were transferred into three porcine uteri. While two pregnant sows were obtained, one sow was unfortunately aborted, ultimately yielding six F0 pigs (Fig. 3a). Of the six pigs, one (#425) showed a target site C-to-G mutation in the GGTA1 gene, which introduced the Q116E mutation in alpha-1,3-galactosyltransferase (Fig. 3b, c; Supplementary Figure 3A), and two (#420, #422) showed a target site C-to-T conversion in the B4galNT2 gene (Fig. 3d, e; Supplementary Figure 3B), and slightly chimeric mutation was found in the CMAH gene of #414 and #425 (Supplementary Figure 3C). Notably, no off-target mutations were detected at the potential off-target sites (POTs) in the pig #420 (Supplementary Figure 4). As shown in Fig. 3f, pig #425, which expressed the Q116E mutation in GGTA1, exhibited diminished alpha-1,3-galactosyltransferase enzyme activity. The indirect immunofluorescence assay (IFA) revealed a significant reduction in cell surface Sd(a) expression in the pig #420 and the pig #422 due to inactivation of beta-1,4-N-acetyl-galactosaminyl transferase 2 compared to that in wild-type pig #414, which was consistent with their genotypes (Fig. 3g).
Fig. 3.
Generating gene mutation pigs by injecting three target sgRNAs and Cas9-BE4-Gam into porcine IVF embryo. a Photo of F0 piglets at 2 weeks after birth. b Sanger sequencing chromatograms of WT and GGTA1 mutation pig (#425). The red arrow indicates the substituted nucleotide. c Product distributions at GGTA1 sites in the pig #425 porcine genome treated with BE4-Gam and the corresponding SgRNA. At each position, 1270332–1270463 sequencing reads were used. d Sanger sequencing results of WT and B4galNT2 mutation pig (#420, #422) at targeted locus. The red arrow indicates the substituted nucleotide. e Product distributions at B4galNT2 sites in the pig #420 porcine genome treated with BE4-Gam and the corresponding SgRNA. At each position, 1561993–1562083 sequencing reads were used. f Detection of aGal expression profiles on the surface of fibroblasts from WT and base mutation pig (#425) by IFA. The nucleus in the PFFs was shown in blue dots and the aGal on the surface of PFFs were shown in red. Bars represent mean ± SD, #425: n = 3, WT: n = 3. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. g Detection of Sd(a) expression profiles on the surface of fibroblasts from WT and base mutation pigs (#420, #422) by IFA. The nucleus in the PFFs is shown in blue dots and the Sd(a) on the surface of PFFs are shown in red. Bars represent mean ± SD, #420, #422: n = 3, WT: n = 3. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001
In summary, we successfully yielded positively mutated pigs by injecting three sgRNAs and CBEs mRNA, although none of the pigs simultaneously exhibited the C-to-T conversion in all three genes. According to these data, we confirmed that this base editor is a simple and efficient method for generating genetically modified pigs.
Efficiently generating three gene mutations in Bama miniature pig genome by somatic cell nuclear transfer (SCNT)
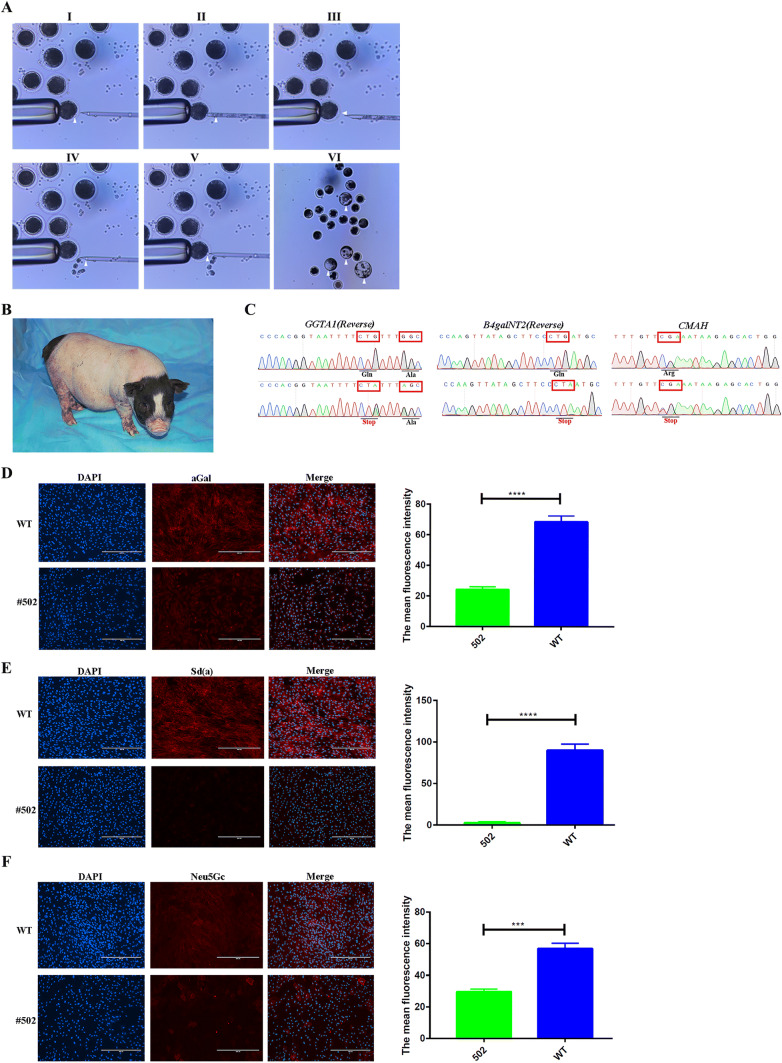
Meanwhile, we used the positive three genes mutated Bama miniature pig fibroblasts clone #53 (described above, shown in Fig. 2d) as donor cells to perform SCNT and the process of SCNT was shown in Fig. 4a. Prior to the reconstructed embryo transfer, wild-type Bama miniature pig fibroblasts and Bama miniature pig fibroblasts clone #53 were used as donor cells to perform SCNT to examine the blastocyst rate of reconstructed embryos. The results shown in Table S1 demonstrated that wild-type Bama miniature pig fibroblasts and Bama miniature pig fibroblasts clone #53 shared a similar blastocyst rate (14.6% ± 0.1% vs 15.8% ± 0.5%, p > 0.05, n = 3), and there was no adverse effect on the development of the reconstructed embryos. Hence, a total of 500 reconstructed embryos were surgically transferred to five recipient uteri, and one was pregnant. After approximately 114 days of gestation, one piglet named #502 who grew and development normally was obtained (Fig. 4b). Notably, Sanger Sequencing showed that this piglet was three genes C-to-T mutated in the genome (Fig. 4c). In addition, there is no off-target mutation at the potential off-target sites were founded (Supplementary Figure 5). Furthermore, the results of IFA revealed that the activity of these three gene-related enzymes in the fibroblasts of #502 are decreased significantly compared to wild-type Bama miniature porcine fibroblasts (Fig. 4d–f). Overall, these data demonstrated that BE4-Gam system introduced three gene mutations in Founder pigs and the potential ability to carry out multiple genes loss-of-functional study.
Fig. 4.
Efficiently generating three gene mutations in Bama miniature pig genome by somatic cell nuclear transfer (SCNT). a Procedure for pig SCNT using Fetal Bama miniature pig fibroblasts. Spindle–chromosome complex (arrowhead) in a porcine MII oocyte before (I) and after (II) removal; an oocyte with a slit in the zona pellucida (III, arrowhead) induced by a beveled glass pipette, and the Fetal Bama miniature pig fibroblasts (arrowhead) before (IV) and after (V) injection into the perivitelline space; the blastocysts developed from injected embryos (VI), which were produced by SCNT using fetal fibroblasts. b Photo of F0 piglet at 4 weeks after birth. c Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and #502. The mutation amino acids are shown in the red boxes. Detection of aGal (d), Sd(a) (e), and Neu5Gc (f) expression profiles on the surface of fibroblasts from WT and base mutation pig (#502) by IFA, respectively. The nucleus in the PFFs is shown in blue dots, and the aGal, Sd(a), and Neu5Gc on the surface of PFFs are shown in red. Bars represent mean ± SD, #502: n = 3, WT: n = 3. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001
Efficiently introducing C-to-T mutations in multiple porcine blastocyst and fibroblast genes using the AncBE4max system
Recently, David Liu’s group developed a new CBE version, termed AncBE4max, which has modified nuclear localization signals (NLSs), modified codon usage, and an ancestrally reconstructed deaminase component. While the high efficiency of base editing has been demonstrated in human HEK293T cells, no related efficiency test has been performed on mammalian embryos; thus, we herein investigated the mutation rates of porcine blastocysts and PFFs induced by the AncBE4max system.
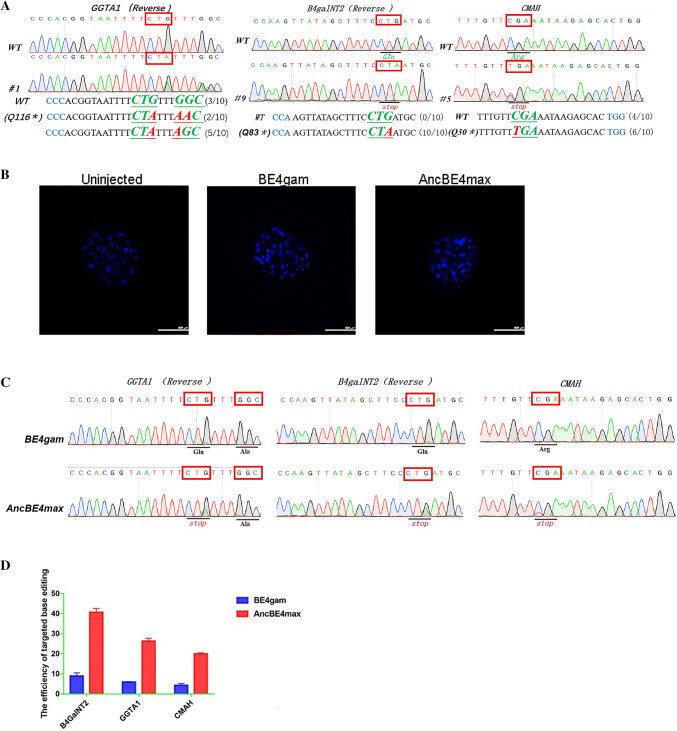
To compare the base-editing efficiencies of AncBE4max and BE4-Gam, we evaluated their mutation rates at the same three target loci, GGTA1, B4galNT2, and CMAH, in the porcine embryo. We simultaneously injected three sgRNAs and AncBE4max mRNA into 100 parthenogenetic embryos, and obtained 14 well-developed blastocysts 7 days later. For the control, we injected three sgRNAs and BE4-Gam mRNA into 110 parthenogenetic embryos. The efficiencies of three genes’ base conversion achieved using the AncBE4max and BE4-Gam systems were 71.4% and 23.1%, respectively; however, an increased ratio of bystander edits, located in the deaminase APOBEC1 deamination window, was also observed when the AncBE4max system was used (Fig. 5a; Table 4; Supplementary Figure 6). Blastocyst development was normal when both systems were used (Fig. 5b).
Fig. 5.
Efficiently multiple genes base editing in porcine parthenogenesis embryo and porcine fibroblasts with AncBE4max. a Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and three gene-edited porcine blastocysts. Target sequence (black), PAM region (blue), target sites (red), and mutant amino acid (bold and italic). b The blastocysts of un-injected blastocysts and the blastocysts which injected three gene sgRNAs plus BE4-Gam or AncBE4max simultaneously. The blastocysts were stained with DAPI, and the nucleus in the blastocyst is shown in blue dots. c Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and Bama miniature PFFs after transfected three gene sgRNAs and BE4-Gam express plasmid. Target amino acid is indicated by red box. d The efficiency comparison of target base conversion between BE4-Gam and AncBE4max
Table 4.
Three genes base-editing rates using AncBE4max or BE4gam system in porcine embryos
| Target genes | Editing system | No. of zygotes | No. of 2-cell (%) | No. of blastocysts (%) | No. of mutants (%) | No. of target mutants (%) | No. of bystander editing (%) | No. of indels (%) | No. of C to non-T (%) | No. of double genes mutants (%) | No. of three genes mutants (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GGTA1 | Anc-BE4max | 100 | 35 (35.0) | 14 (14.0) | 10/14 (71.4) | 9/14 (64.2) | 10/14 (71.4 ) | 0/14 (0) | 1/14 (7.14) | 0/14 (0) | 10/14 (71.4) |
| B4GalNT2 | 10/14 (71.4) | 10/14 (71.4) | 4/14 (28.5) | 0/14 (0) | 0/14 (0) | ||||||
| CMAH | 14/14 (100) | 14/14 (100) | 0/14 (0) | 0/14 (0) | 0/14 (0) | ||||||
| GGTA1P | BE4-Gam | 110 | 37 (33.7) | 13 (11.8) | 6/13 (46.2) | 6/13 (46.2) | 3/13 (23.1) | 0/13 (0) | 1/13 (7.7) | 2/13 (15.4) | 3/13 (23.1) |
| B4GalNT2 | 6/13 (46.2) | 6/13 (46.2) | 1/13 (7.7) | 0/13 (0) | 0/13 (0) | ||||||
| CMAH | 5/13 (38.5) | 5/13 (38.5) | 0/13 (0) | 0/13 (0) | 0/13 (0) |
Similarly, we transfected these three sgRNAs and BE4-Gam or AncBE4max expression plasmids into Bama miniature PFFs. Three days later, DNA was extracted from these fibroblasts and sequenced. Consistent with the blastocyst results, the efficiency of the AncBE4max system to convert bases in the PFF genome was four-to-fivefold higher than that of the BE4-Gam system (Fig. 5c, d). Furthermore, we took the three genes mutational PFFs named Anc-18 (Supplementary Figure 7A) which was obtained at 12 days later after transfecting three sgRNAs and AncBE4max expression plasmids to compare the precision at the POTs with that of BE4-Gam. The result (Supplementary Figure 7B–7D) indicated that there is no off-target event which was found which imply that this AncBE4max system shares the high efficiency and precision.
Taken together, these results demonstrated that AncBE4max edited bases at a high rate but simultaneously produced more unwanted mutations; thus, this method will significantly enhance the efficiency of porcine genome editing.
Discussion
To our knowledge, our study is the first report of using BE4-Gam and AncBE4max to precisely introduce three gene base conversions into the porcine genome, although there is a similar report using BE3 to introduce single base mutation in pig [19]. Our data revealed that BE4-Gam can induce precise base substitutions at target sites with efficiencies of 66.7–71.4% and 5.3–10.1% in porcine blastocysts and Bama miniature pig PFFs, respectively, upon the injection of BE4-Gam mRNA and associated single sgRNA. Furthermore, simultaneously introducing three sgRNAs targeting the GGTA1, B4galNT2, and CMAH genes and BE4-Gam into porcine parthenogenic embryos or PFFs resulted in the mutation all three targeted bases at a rate of 18.1% in the blastocyst genome and 7.5% in the fibroblast genome. In addition, in porcine blastocysts, the newly developed BE version (AncBE4max) exhibited a three gene base conversion efficiency (71.4%) better than that of the control BE4-Gam system (approximately 23.1%) (Table 4). The three gene base conversion efficiency of AncBE4max was four-to-fivefold higher than that of BE4-Gam in PFFs. These results indicated that CBEs can simply and efficiently edit targeted bases in both porcine embryos and fibroblasts.
To date, most animal models have been created using wild-type (WT) Cas9 to introduce DSBs in DNA, which has raised some concerns. First, not all indels result in gene silencing, and some may even cause the targeted protein to have new functions [26]. Second, this method may induce excessive DNA damage and reduce fitness when high copy-number regions are targeted [50–53]. Third, the Cas9 method cannot efficiently be used to explore the effects of specific point mutations within genes to make comparisons between alleles [54] or precisely mimic human diseases [4]. Most importantly, the majority of known human genetic diseases are induced by point mutations, which are also known as SNPs [18, 20]. The mainstream approach to introducing a desired base mutation in the genomes of porcine [2] and other animals [55] is mediated by CRISPR/Cas9 systems using single-stranded oligodeoxyribonucleotide (ssODNs) or long ssDNAs via homology-directed repair (HDR), but some technological challenges remain due to the high frequency of random indels and low efficiency of base substitutions [16, 17]. In contrast, base editors, especially AncBE4max systems, more simply and efficiently edit target bases in porcine embryos and fibroblasts, as these editors require no DSBs or exogenous DNA templates and introduce random indels at a low frequency. Furthermore, there are no reports of using the CRISPR/Cas9 system with ssODNs to introduce multiple gene point mutations in animal embryos or cells in only one step via HDR. This study is the first to successfully introduce three gene mutations in porcine blastocysts and PFFs using CBEs in only one step, and produce three xeno-antigen-related gene knock out pig by SCNT. By injecting the complex of sgRNAs and BE4-Gam mRNA into IVF embryo, we also report precise base conversion within the porcine B4galNT2 gene to inactive beta-1,4-N-acetyl-galactosaminyl transferase 2, and obtained a pig with the GGTA1 Q116E (C to G) mutation which has been observed in porcine blastocysts, PFFs, and other animals [4] at a low frequency. The alpha-1,3-galactose (alpha-gal), the cell surface glycan antigen Sd(a) and the cell surface molecule sialic acid Neu5Gc expression in the PFF isolated from the pig with a positive C to T mutation in the GGTA1, B4galNT2 and CMAH gene were significantly decreased, and the pig with the C-to-G mutation in the GGTA1 gene exhibited decreased alpha-1,3-galactosyltransferase activity. Unfortunately, no pigs were positive for the C to T in the GGTA1 gene, the B4galNT2gene, and CMAH gene simultaneously which was obtained after transplant the IVF embryos into recipient uterus. We speculate that this result may have been due to the low efficiency of IVF and the polyspermy phenomenon, which impacts the implantation and development of blastocysts and leads to fewer piglets. Furthermore, embryonic analysis after the in vitro injection of three sgRNAs and BE4-Gam mRNA into porcine parthenogenetic embryos demonstrated a simultaneous three gene mutations efficiency of only approximately 18.1–23.1%, which may be another factor underlying the failure to generate three gene-mutated pigs. We acknowledge that, together, these factors may have accounted for our failure to generate pigs with three gene mutations. Consistent with previous reports [4, 20, 56], no off-target events occurred in the POTs of the base-edited pigs. Although a recently report [57] showed that the BE3 off-target SNVs were induced by overexpression of APOBEC1 and were sgRNA-independent in mouse embryos, it needs further experiment to explore the precision of BE4-Gam or the others CBEs in the region outside the POTs of porcine embryo genome.
We also tested the efficiency of AncBE4max by injecting three sgRNAs and AncBE4max into porcine parthenogenetic embryos in vitro, revealing that AncBE4max was more efficient than the previous version (BE4-Gam), as it yielded a simultaneous three gene mutations rate of 71.4%; thus, AncBE4max may play a more important role in generating multiple gene mutation animal models in the future. At the same times, this AncBE4max system shares the same precision at the POTs with the BE4-Gam system. Nevertheless, the ratio of bystander edits induced by AncBE4max was obviously increased compared with that generated by BE4-Gam, which may have been due to improvement in the expression of soluble proteins and the high activity of APOBEC1. Although bystander editing may not be especially important when using base editing to disrupt promoters, splice sites, or other regulatory elements or when silencing genes by introducing premature stop codons, it may affect the production of animal models that precisely mimic human disorders. Bystander editing may be minimized by combining the BE architecture of AncBE4max with optimized mutations in the rAPOBEC1 domain or the engineered human APOBEC3A (eA3A) domain. Therefore, this base editor may be a reliable tool for editing bases with high efficiency, precision, and specificity.
In summary, we are the first to edit bases in a programmable manner using BE4-Gam in pigs, and upgrading to AncBE4max significantly increased the base-editing efficiency in porcine blastocysts and PFFs; albeit this method did increase the frequency of bystander edits. Thus, CBEs are simple and efficient tools for editing bases in porcine embryos and fibroblasts. Furthermore, our results suggest that CBEs have enormous potential for improving the economic traits, breeding cycle, and disease tolerance in pigs, such that they can be used to mimic human diseases.
Materials and methods
Ethics statement
All animal studies were approved by the Animal Welfare and Research Ethics Committee of Jilin University, and all procedures were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals. All pigs were obtained from the Huichang Animal Husbandry Science and Technology Co., Ltd. All surgeries were performed under anesthesia, and every effort was made to minimize animal suffering.
Vector construction
SgRNA oligonucleotides targeting the GGTA1, B4galNT2, and CMAH genes, constructed using online software, were annealed and cloned into the puc57-sgRNA or pBluescriptSKII-U6-sgRNA expression vectors. The detailed sequences are provided in Fig. 1a.
mRNA and RNA preparation
The BE4-Gam and AncBE4max plasmids were purchased from Addgene (#100806, #112094). These plasmids were linearized with Not I, and mRNAs were synthesized with an in vitro RNA transcription kit (HiScribe™ T7 ARCA mRNA kit (with tailing), NEB). sgRNAs were amplified and transcribed in vitro using the MAXIscript T7 kit (Ambion) and purified using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions.
Microinjection of porcine zygotes
As shown in our published protocols, a mixture of BE4-Gam or AncBE4max mRNA (200 ng/μl) and corresponding sgRNAs (50 ng/μl) was injected into the cytoplasms of porcine parthenogenetic embryos or IVF embryos.
Cell culture and transfection
Porcine fibroblasts were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10% fetal bovine serum. Approximately 3 × 106 PFFs were electroporated with 25 μg of the BE4-Gam vector or the AncBE4max vector together with 20 μg of the sgRNA expression plasmid in 250 μl of Opti-MEM (Gibco) using BTX ECM 2001 (Harvard Bioscience, Holliston, MA, USA). The electroporation parameters for 2 mm gap cuvettes were as follows: 340 V, 1 ms, and 3 pulses for 1 repeat.
Genotyping of PFF clones and single embryos
The cells were plated into 100 mm dishes at 72 h post-transfection at an appropriate density; the cell inoculation density was 2500 cells/per 100 mm dish on average. The cell clones were picked and cultured in 24-well plates. Approximately 15% of each cell clone or single embryo was digested and lysed with 10 μl of NP40 lysis buffer (0.45% NP40 plus 0.6% proteinase K) for 1 h at 56 °C and 10 min at 95 °C to provide templates for genotyping. The primers for genotyping are shown as follows: GGTA1 (forward 5′-GTGGTATGGGAAGGCA-CT-3′; reverse 5′-GACCTCAGCAGAAGGGAG-3′); B4GalNT2 (forward 5′-ACCA-GCCCACTTTCCCAATA-3′; reverse 5′-CGCAAGAGCCCTCAGCAT-3′); CMAH (forward 5′-GCAGCATCGAACAAACGAC-3′, reverse 5′-GCACATTTCCTGCCA-AAC-3′).
Somatic cell nuclear transfer (SCNT)
The protocol used for SCNT was performed as described by Lai et al. [58]. In summary, as shown in Fig. 4, matured oocytes were collected and enucleated using a beveled glass pipette by aspirating the first polar body and the metaphase II plate in a small amount of surrounding cytoplasm in a manipulation medium of HEPES-buffered M199 plus cytochalasin B. Then the PFFs cells were injected into the perivitelline cytoplasm of enucleated oocytes using the same slit in the zona pellucida as made during enucleation. Finally, two DC pulses at 1.2 kV/cm for 30 μs using an electrofusion instrument successively fused and activated the produced embryos. The reconstructed embryos were transferred to the oviducts of surrogates after overnight culture in PZM3 at 39 °C.
Detection of the mutation in the cloned pigs
To confirm the piglet genotypes, genomic DNA extracted from the ears of cloned piglets was used as a template for PCR using the primer pairs described in Supplementary Data 1.
Targeted deep sequencing
Targeted sites were amplified from genomic DNA using Phusion polymerase (Thermo Fisher Scientific). The paired-end sequencing of PCR amplicons was performed by Novogene Bioinformatics Technology Co. Ltd (Beijing), using an Illumina MiSeq.
Off-target assay
The potential off-target sites (POTs) for each sgRNA were predicted to analyze site-specific edits according to Cas-OFFinder (http://www.rgenome.net/cas-offinder/). In summary, three bases mismatch and 0 DNA or RNA bulge are accepted for the off-target assay. The PCR products of the POTs were sequenced. All primers for the off-target assay are provided in Supplementary Data 2.
The isolation of PFFs
Porcine fibroblasts were derived from the tails of seven piglets as follows: the tails from seven piglets were disaggregated in culture medium containing 200 U/ml collagenase IV (type IV, 260 U/mg, Gibco, Grand Island, NY, USA), 0.0125 mg/ml DNase I (2000 U/mg, Sigma, St. Louis, MO, USA), 20% fetal bovine serum (Gibco, Grand Island, NY, USA), and 1% penicillin/streptomycin (Gibco) for 4–6 h. Isolated PFFs were resuspended and cultured in 10-cm cell culture dishes until they reached sub-confluence. Cells at passage 1 were frozen in fetal bovine serum containing 10% dimethylsulfoxide.
Indirect immunofluorescence assay (IFA)
Three well-characterized xeno-antigen have been implicated, including α-Gal, Sd(a), and N-glycolylneuraminic acid whose synthesis are catalyzed by the GGTA1 gene, the B4GalNT2 gene, and the CMAH gene-related enzyme, respectively. To detect the α-Gal antigen, PFFs were stained with biotin-conjugated GSI-B4 (1938420, Invitrogen) (1:200) at 37 °C for 1 h, and then incubated with Streptavidin-PE (1992345, Invitrogen) (1:100) at 37 °C for 0.5 h. For the Sd(a) antigen, PFFs were stained with biotin-conjugated Dolichos biflorus agglutinin (DBA) (B-1035, Vector Laboratories) (1:200) at 37 °C for 1 h, and then incubated with Streptavidin-PE (1992345, Invitrogen) (1:100) at 37 °C for 0.5 h. To detect the N-glycolylneuraminic acid, PFFs were stained with Anti Neu5Gc Antibody (146903, Biolegend) (1:250) at 37 °C for 1 h, and then incubated with R-Phycoerythrin+-conjugated Donkey Anti-Chicken IgY++ (125844, Jackson ImmunoResearch) (1:200) at 37 °C for 0.5 h. 4,6-Diamidino-2-phenylindole (DAPI, Invitrogen) (1:1000) was applied for nuclear staining. Then, the results were assessed by fluorescence microscopy (Olympus BX51). The fluorescence intensity was quantified using image processing (ImageJ, NIH, Bethesda, MD, USA) on unmanipulated TIFF images.
Statistic
Statistical significance was calculated using two-tailed unpaired Student’s t test (Figs. 3f, g, 4c–e).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1 Targeted base conversion in porcine blastocysts with BE4-Gam by injecting single sgRNA. A-C The DNA fragments of GGTA1 (A), B4galNT2 (B) and CMAH (C) from the porcine blastocysts were sub-cloned into pGM-T vectors and then sequenced, respectively. The number of clones for each sequence pattern is indicated. The targeted sequence was underlined, the PAM sequence was shown with green and substituted nucleotide was shown with red. (TIFF 10622 kb)
Supplementary Figure 2 Targeted base conversion in porcine blastocysts with BE4-Gam by injecting three gene sgRNAs. A-C DNA fragments of GGTA1 (A), B4galNT2 (B) and CMAH (C) from the porcine blastocysts were sub-cloned into pGM-T vectors and sequenced, respectively. The number of clones for each sequence pattern is indicated. The targeted sequence was underlined, the PAM sequence was shown in green and substituted nucleotide was shown in red. (TIFF 15896 kb)
Supplementary Figure 3 The results of deep sequencing from six IVF derived pigs. A-C The results of deep sequencing of GGTA1 (A), B4galNT2 (B) and CMAH (C)genes from all IVF derived F0 pigs, respectively. At each position, 1170407-1562083 sequencing reads were used. (TIFF 9229 kb)
Supplementary Figure 4 Off-target detection in the pig # 420. Chromatogram sequence analysis of potential off-target sites (POTS) for sgRNA in the pig #420 genome of GGTA1 (A), B4galNT2 (B), CMAH (C), respectively. 20 bp of the POTS and the PAM are represented in shadow. (TIFF 2333 kb)
Supplementary Figure 5 Off-target detection in the pig # 502. Chromatogram sequence analysis of potential off-target sites (POTS) for sgRNA in the pig #502 genome of GGTA1 (A), B4galNT2 (B), CMAH (C), respectively. 20 bp of the POTS and the PAM are represented in shadow. (TIFF 10862 kb)
Supplementary Figure 6 Efficiently multiple genes base editing in porcine parthenogenesis embryo with AncBE4max. A-F The genotypes of GGTA1, B4galNT2 and CMAH mutant blastocysts which injected AncBE4max and three gene sgRNAs, shown in A, C and E, respectively. The genotypes of GGTA1, B4galNT2 and CMAH mutant blastocysts which injected BE4-Gam and three gene sgRNAs, shown in B, D, F respectively. The number of clones for each sequence pattern is indicated. Target sequence (underlined), PAM region (green), and substituted nucleotide (red). (TIFF 20544 kb)
Supplementary Figure 7 Off-target detection in the Anc-18 PFFs. A Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and Bama miniature Anc-18 PFFs after transfected three gene sgRNAs and Anc-BE4max express plasmid. Target amino acid are indicated by red box. B-D Chromatogram sequence analysis of potential off-target sites (POTS) for sgRNA in the pig #502 genome of GGTA1 (B), B4galNT2 (C), CMAH (D), respectively. 20 bp of the POTS and the PAM are represented in shadow. (TIFF 14256 kb)
Acknowledgements
The authors thank Zhuang Shao, Chuang Gao, and Kang Yang for assistance at the Embryo Engineering Center for the critical technical assistance.
Author contributions
Conceived and designed the experiments: HO and DP. Performed the experiments: HY, TY, LW, LY, YZ, HL, ML, XT, ZL, ZL, CL, and XC. Wrote the manuscript: HO and DP. All authors reviewed the manuscript.
Funding
This work was supported by Special Funds for Cultivation and Breeding of New Transgenic Organisms (No. 2016ZX08006001), the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, No. IRT_16R32), the Program for JLU Science and Technology Innovative Research Team (2017TD-28), and the Fundamental Research Funds for the Central Universities.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongming Yuan, Tingting Yu, and Lingyu Wang have contributed equally to this work.
Contributor Information
Daxin Pang, Email: pdx@jlu.edu.cn.
Hongsheng Ouyang, Email: ouyh@jlu.edu.cn.
References
- 1.Li M, et al. Site-specific fat-1 knock-in enables significant decrease of n-6PUFAs/n-3PUFAs ratio in pigs. G3 (Bethesda) 2018;8(5):1747–1754. doi: 10.1534/g3.118.200114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K, et al. Efficient generation of myostatin mutations in pigs using the CRISPR/Cas9 system. Sci Rep. 2015;5:16623. doi: 10.1038/srep16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K, et al. Efficient generation of orthologous point mutations in pigs via CRISPR-assisted ssODN-mediated homology-directed repair. Mol Ther Nucleic Acids. 2016;5(11):e396. doi: 10.1038/mtna.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, et al. Highly efficient RNA-guided base editing in rabbit. Nat Commun. 2018;9(1):2717. doi: 10.1038/s41467-018-05232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, et al. CRISPR/Cas9-mediated GJA8 knockout in rabbits recapitulates human congenital cataracts. Sci Rep. 2016;6:22024. doi: 10.1038/srep22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sui T, et al. CRISPR/Cas9-mediated mutation of PHEX in rabbit recapitulates human X-linked hypophosphatemia (XLH) Hum Mol Genet. 2016;25(13):2661. doi: 10.1093/hmg/ddw125. [DOI] [PubMed] [Google Scholar]
- 7.Zou Q, et al. Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol. 2015;7(6):580. doi: 10.1093/jmcb/mjv061. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinberger G, et al. The FTD-like syndrome causing TREM2 T66M mutation impairs microglia function, brain perfusion, and glucose metabolism. EMBO J. 2017;36(13):1837–1853. doi: 10.15252/embj.201796516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Zhu JK. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant. 2017;10(3):523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Zong Y, et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):438. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 12.Gu T, et al. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci. 2018;9:3248–3253. doi: 10.1039/c8sc00637g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilton IB, Gersbach CA. Enabling functional genomics with genome engineering. Genome Res. 2015;25(10):1442–1455. doi: 10.1101/gr.190124.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;Database issue(1):D980. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koblan LW, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox DBT, Randall Jeffrey P, Feng Z. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y, et al. Correction of the Marfan syndrome pathogenic FBN1 mutation by base editing in human cells and heterozygous embryos. Mol Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, et al. Efficient RNA-guided base editing for disease modeling in pigs. Cell Discov. 2018;4:64. doi: 10.1038/s41421-018-0065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K, et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35(5):435. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Programmable base editing of zebrafish genome using a modified CRISPR–Cas9 system. Nat Commun. 2017;8(1):118. doi: 10.1038/s41467-017-00175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimatani Z, et al. Targeted base editing in rice and tomato using a CRISPR–Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):441. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 23.Ren B, et al. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci China Life Sci. 2017;60(5):516–519. doi: 10.1007/s11427-016-0406-x. [DOI] [PubMed] [Google Scholar]
- 24.Yuan J, et al. Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase. Mol Cell. 2018;72(2):380–394.e7. doi: 10.1016/j.molcel.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Gapinske M, et al. CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol. 2018;19(1):107. doi: 10.1186/s13059-018-1482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuscu C, et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14(7):710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, et al. Generation of GTKO Diannan miniature pig expressing human complementary regulator proteins hCD55 and hCD59 via T2A peptide-based bicistronic vectors and SCNT. Mol Biotechnol. 2018;60(8):550–562. doi: 10.1007/s12033-018-0091-6. [DOI] [PubMed] [Google Scholar]
- 28.Lai L, et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol. 2006;24(4):435. doi: 10.1038/nbt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naruse K, et al. Production of a transgenic pig expressing human albumin and enhanced green fluorescent protein. J Reprod Dev. 2005;51(4):539. doi: 10.1262/jrd.16073. [DOI] [PubMed] [Google Scholar]
- 30.Peng J, et al. Production of human albumin in pigs through CRISPR/Cas9-mediated knockin of human cDNA into swine albumin locus in the zygotes. Sci Rep. 2015;5:16705. doi: 10.1038/srep16705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren Z, et al. Enhancement of porcine intramuscular fat content by overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase in skeletal muscle. Sci Rep. 2017;7:43746. doi: 10.1038/srep43746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Draghiaakli R, et al. Myogenic expression of an injectable protease-resistant growth hormone–releasing hormone augments long-term growth in pigs. Nat Biotechnol. 1999;17(12):1179. doi: 10.1038/70718. [DOI] [PubMed] [Google Scholar]
- 33.Burkard C, et al. Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR34 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017;13(2):e1006206. doi: 10.1371/journal.ppat.1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prather RS, et al. Knockout of maternal CD163 protects fetuses from infection with porcine reproductive and respiratory syndrome virus (PRRSV) Sci Rep. 2017;7(1):13371. doi: 10.1038/s41598-017-13794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells KD, et al. Substitution of porcine CD163 SRCR domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome (PRRS) viruses. J Virol. 2016 doi: 10.1128/JVI.01521-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitworth KM, Prather RS. Gene editing as applied to prevention of reproductive porcine reproductive and respiratory syndrome. Mol Reprod Dev. 2017;84(9):926–933. doi: 10.1002/mrd.22811. [DOI] [PubMed] [Google Scholar]
- 37.Yang H, et al. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antivir Res. 2018;151:63–70. doi: 10.1016/j.antiviral.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Niu D, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR–Cas9. Science. 2017;357(6357):1303. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estrada JL, et al. Evaluation of human and non-human primate antibody binding to pig cells lacking GGTA1/CMAH/β4GalNT2 genes. Xenotransplantation. 2015;22(3):194–202. doi: 10.1111/xen.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan S, et al. A Huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington’s disease. Cell. 2018;173(4):989–1002.e13. doi: 10.1016/j.cell.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, et al. Reducing immunoreactivity of porcine bioprosthetic heart valves by genetically-deleting three major glycan antigens, GGTA1/β4GalNT2/CMAH. Acta Biomater. 2018;72:196–205. doi: 10.1016/j.actbio.2018.03.055. [DOI] [PubMed] [Google Scholar]
- 42.Cullot G, et al. CRISPR–Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. 2019;10(1):1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo E, et al. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017;18(1):224. doi: 10.1186/s13059-017-1354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adikusuma F, et al. Large deletions induced by Cas9 cleavage. Nature. 2018;560(7717):E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 45.Hysi PG, et al. Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nat Genet. 2018;50(5):652–656. doi: 10.1038/s41588-018-0100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin SC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49(7):1152. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullin S, Schapira A. The genetics of Parkinson’s disease. Br Med Bull. 2015;114(1):292–298. doi: 10.1093/bmb/ldv022. [DOI] [PubMed] [Google Scholar]
- 49.Noh HJ, et al. Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat Commun. 2017;8(1):774. doi: 10.1038/s41467-017-00831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163(6):1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 51.Wang T, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168(5):890–903.e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguirre AJ, et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016;6(8):914. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ihry RJ, et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 54.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inui M, et al. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep. 2014;4:5396. doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D, et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat Biotechnol. 2017;35(8):475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- 57.Zuo E, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364(6437):289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan N, et al. Piglets cloned from induced pluripotent stem cells. Cell Res. 2013;23(1):162–166. doi: 10.1038/cr.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Targeted base conversion in porcine blastocysts with BE4-Gam by injecting single sgRNA. A-C The DNA fragments of GGTA1 (A), B4galNT2 (B) and CMAH (C) from the porcine blastocysts were sub-cloned into pGM-T vectors and then sequenced, respectively. The number of clones for each sequence pattern is indicated. The targeted sequence was underlined, the PAM sequence was shown with green and substituted nucleotide was shown with red. (TIFF 10622 kb)
Supplementary Figure 2 Targeted base conversion in porcine blastocysts with BE4-Gam by injecting three gene sgRNAs. A-C DNA fragments of GGTA1 (A), B4galNT2 (B) and CMAH (C) from the porcine blastocysts were sub-cloned into pGM-T vectors and sequenced, respectively. The number of clones for each sequence pattern is indicated. The targeted sequence was underlined, the PAM sequence was shown in green and substituted nucleotide was shown in red. (TIFF 15896 kb)
Supplementary Figure 3 The results of deep sequencing from six IVF derived pigs. A-C The results of deep sequencing of GGTA1 (A), B4galNT2 (B) and CMAH (C)genes from all IVF derived F0 pigs, respectively. At each position, 1170407-1562083 sequencing reads were used. (TIFF 9229 kb)
Supplementary Figure 4 Off-target detection in the pig # 420. Chromatogram sequence analysis of potential off-target sites (POTS) for sgRNA in the pig #420 genome of GGTA1 (A), B4galNT2 (B), CMAH (C), respectively. 20 bp of the POTS and the PAM are represented in shadow. (TIFF 2333 kb)
Supplementary Figure 5 Off-target detection in the pig # 502. Chromatogram sequence analysis of potential off-target sites (POTS) for sgRNA in the pig #502 genome of GGTA1 (A), B4galNT2 (B), CMAH (C), respectively. 20 bp of the POTS and the PAM are represented in shadow. (TIFF 10862 kb)
Supplementary Figure 6 Efficiently multiple genes base editing in porcine parthenogenesis embryo with AncBE4max. A-F The genotypes of GGTA1, B4galNT2 and CMAH mutant blastocysts which injected AncBE4max and three gene sgRNAs, shown in A, C and E, respectively. The genotypes of GGTA1, B4galNT2 and CMAH mutant blastocysts which injected BE4-Gam and three gene sgRNAs, shown in B, D, F respectively. The number of clones for each sequence pattern is indicated. Target sequence (underlined), PAM region (green), and substituted nucleotide (red). (TIFF 20544 kb)
Supplementary Figure 7 Off-target detection in the Anc-18 PFFs. A Representative sequencing chromatograms at the GGTA1, B4galNT2, and CMAH targets of WT and Bama miniature Anc-18 PFFs after transfected three gene sgRNAs and Anc-BE4max express plasmid. Target amino acid are indicated by red box. B-D Chromatogram sequence analysis of potential off-target sites (POTS) for sgRNA in the pig #502 genome of GGTA1 (B), B4galNT2 (C), CMAH (D), respectively. 20 bp of the POTS and the PAM are represented in shadow. (TIFF 14256 kb)