Abstract
Maintenance of genome stability is a crucial cellular function for normal mammalian development and physiology. However, despite the general relevance of this process, genome stability alteration due to genetic or non-genetic conditions has a particularly profound impact on the developing cerebral cortex. In this review, we will analyze the main pathways involved in maintenance of genome stability, the consequences of their alterations with regard to central nervous system development, as well as the possible molecular and cellular basis of this specificity.
Keywords: DNA damage, Chromosome stability, Microcephaly, MCPH, BER, NER, Fanconi anemia, SCKS, HR, NHEJ, Mitosis, Centrosome
Introduction
Maintenance of genome stability is a complex cellular task, handled by several interrelated mechanisms that ensure faithful duplication and segregation of genetic material during cell cycle, as well as its repair in proliferating or post-mitotic cells. Alterations of these mechanisms, due to genetic or non-genetic conditions, have severe consequences, such as cancerous transformation or altered embryonic development. Among the different tissues, developing human neocortex appears to be particularly vulnerable to alterations of genome stability, as one of the most common clinical manifestations of these disorders is microcephaly, which is the result of brain volume reduction. Microcephaly is a common clinical condition that may be evident at birth (primary or congenital microcephaly) or postnatally (secondary or progressive microcephaly). In the hereditary forms, microcephaly may be the only defect observed (non-syndromic microcephaly or MCPH) or be associated with other malformations (syndromic microcephaly) [1]. Dissecting the molecular and cellular mechanisms implicated in genetic primary microcephaly could also be critical to better understand non-genetic forms, such as intrauterine insults, viral infections [2, 3], or alcohol/drug abuse during pregnancy [4]. In theory, any condition that alters neural progenitor expansion or their survival may induce microcephaly. However, independently of the specific alteration of brain development that characterize microcephaly syndromes, the identification of causal mutations has revealed that they mostly occur in ubiquitously expressed genes, whose products are tightly involved in cell proliferation and maintenance of genomic stability. Understanding the reasons of this specificity is in most cases an open issue. In this review, we will summarize the main cellular events in the development of the cerebral cortex, with particular emphasis on those that may distinguish cortical progenitors from other developing cell types. We will also underscore the evidence of specific sensitivity of neural progenitors to genome integrity. Then, we will analyze the genetic causes of microcephaly, grouping the different syndromes on the basis of the main molecular process involved. Finally, we will try to highlight how the interaction between different mechanisms may represent a common framework at the basis of the selective vulnerability of neural progenitors.
Milestone events in cortical development
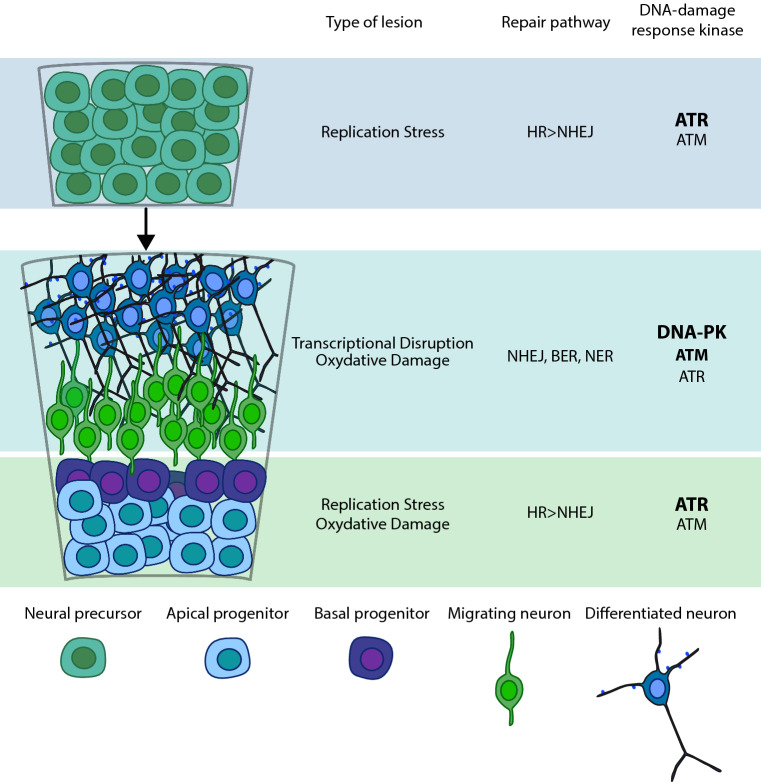
In vertebrates, the earliest stages of brain development require the segmentation of neural tube along a head-to-tail (anteroposterior) and back-to-front (dorsoventral) axes [5]. The first population of neural progenitors consists of neuroepithelial cells (NECs), which expand their pool by undergoing a series of symmetric proliferative divisions [6]. Then, NECs become more fate-restricted radial glial cells (RGCs). RGCs are bipolar cells that retain an apical–basal orientation and express astroglial markers [7, 8]. RGCs contact the pial surface and the ventricular surface trough their basal and apical process, respectively. The onset of neurogenesis is determined by asymmetric division of RGCs cells, i.e., divisions that generate two different daughter cells. RGC asymmetric divisions can be directly neurogenic when they produce an RGC and a neuron or may generate an RGC and a more fate-restricted, Basal Progenitor (BP). BPs undergo one or few rounds of symmetric divisions before giving rise to post-mitotic neuroblasts and are one of the major sources of cortical neurons [9]. RGCs may also divide symmetrically to generate two BPs or two neurons [10]. Different daughter cells are spatially segregated: RGCs cell bodies are restricted to the more apical part of the cortical wall, corresponding to the ventricular zone (VZ); BPs bodies form a more basal layer known as the subventricular zone (SVZ). Importantly, additional progenitor populations have been described, such as the basal RG-like cells [11], which are located abventricularly and are found abundantly in brains of gyrencephalic species, such as human, monkey, and ferret. Newborn cortical neurons migrate along the intermediate zone (IZ) to reach the cortical plate (CP), where they accumulate below the basal lamina. Later-born neurons arriving in the CP migrate past earlier born neurons in an inside-out fashion, thus generating multi-layered neocortex [11] (Fig. 1).
Fig. 1.
Multiple kinds of genome lesions can impact the nervous system at all stages of development and maturity. Replication stress primarily affects proliferating neural progenitors that may require DNA DSB repair, which involves either HR or NHEJ. HR is dependent on using replicated sister chromatids as an error-free repair templates and is not active in non-replicating or differentiated cells. In this phase, ATR exerts a prominent role in protecting cells from DNA damage arising during S phase in consequence of DNA stressors. Replicating cells may also experience other types of damage, including inter-strand crosslinks or strand breaks from other sources, including oxidative lesions and transcription-associated damage. ATM and DNA-PK are activated by DNA DSBs and exert a pivotal function in DSB recognition. In non-cycling cells, NHEJ repairs DNA DSBs, whereas other types of DNA damage require the BER/single-stranded break repair (SSBR) pathway or NER. Oxidative damage can also impact immature cells, but will be an ongoing threat to the mature nervous system
General impact of genomic stability on brain development and function
Genomic instability (GIN) is defined by an increased mutation rate, due to inactivation of DNA repair pathways or to increase of genotoxic stress from cellular processes that overcome high-fidelity DNA repair. Chromosomal instability (CIN) is a subset of GIN, characterized by altered stability of chromosomes, leading to amplification, deletion, or rearrangement of long chromosomal fragments [12, 13]. GIN may result from abnormalities in different steps of cell cycle, including DNA replication [14], abnormal chromosome segregation [15], or abortive cytokinesis [16]. CIN and GIN are especially harmful to cells that continue to proliferate, as they are able to deregulate cell division and prompt senescence, cell death, or uncontrolled proliferation. However, CIN and GIN can also be detrimental in long-lived post-mitotic cells, since they may alter gene expression programs and lead to the production of abnormal proteins [17]. On this basis, it is not surprising that genetic and non-genetic alterations leading to CIN and GIN have profound consequences on CNS development, which is characterized by explosive waves of cell proliferation and generates cells with the longest lifespan in the body. Indeed, CNS is consistently affected in most conditions resulting in CIN and GIN. Interestingly, although these conditions should generically affect all proliferating cells in the body, in many cases, the severity of CNS alterations is disproportionate if compared to other tissues, indicating that CNS development and function have specific vulnerability factors. In particular, neural progenitors show strong sensitivity to DNA damage [18], which may change at different developmental stages [19] (Fig. 2).
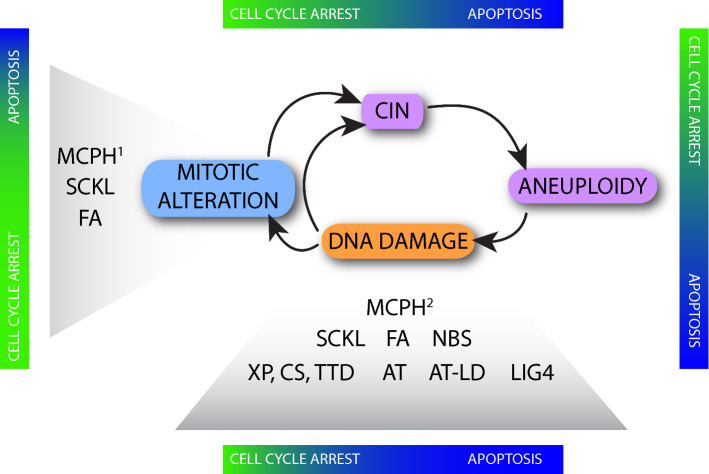
Fig. 2.
Cellular defects that impair the fidelity of mitosis promote chromosome mis-segregation (chromosome instability, CIN) and aneuploidy. Increasing evidence reveals that errors in mitosis can also promote the direct and/or indirect acquisition of DNA damage and chromosome breaks. Unrepaired DNA damage is also deemed a contributor to aneuploidy, which might be especially relevant in the central nervous system (CNS) due to evidence for a low repair capacity of neuronal precursors and differentiated neurons. The induction of DNA damage may lead to apoptosis, which occurs in the majority of conditions described in this review. In contrast, some MCPH, Seckel syndrome and FA are characterized by cell-cycle arrest due to mitotic alterations. Color-coded bars indicate the relative contribution to the cortical phenotype from cell-cycle arrest (green) and apoptosis (blue). MCPH1: all the genes mutated in MCPH that show mitotic alterations. MCPH2: MCPH1, ASPM, and CITK
One of the most dramatic evidences of this sensitivity has been provided by follow-up studies of atomic bomb survivors in Hiroshima and Nagasaki, showing that the frequency of microcephaly in newborns exposed to radiations in utero increased ten times more than the incidence of radiation-induced tumors [20]. In addition, a huge array of human syndromes characterized by neurodevelopmental defects, neurodegeneration, or brain tumors displays defects in DNA-damage signaling [21].
This peculiar sensitivity has been correlated with the relatively low capability of different neural progenitors to cope with DNA damage. It has been shown that neuronal progenitors process IR-induced DNA damage more slowly than neurons and, as a consequence, are also more susceptible to IR-induced apoptosis [22, 23]. A second factor that may justify the specific susceptibility of different neural progenitors to genotoxic insults is the different durations of their cell-cycle phases. G1-phase lengthening is associated with differentiating progenitors [24]: neural progenitors undergoing rapid expansion have longer S phase, compared with progenitors committed to the neurogenic lineage [25]. Longer replicative phase during early progenitor expansion may facilitate accurate DNA repair through homologous recombination (HRR), which requires more time to be completed than non-homologous end joining (NHEJ) [26], thus avoiding transmission of genetic errors to a large population of brain cells. On the other hand, differentiating progenitors rely more on NHEJ to repair DSBs [27], which can be more effective for coping with the high replicative stress associated with rapid proliferation of late neural progenitors [28–30]. Most knock out mouse models of DNA repair genes, such as Polb and Rev7 mutants [31, 32], show lethal phenotypes and neuronal defects associated with apoptosis of neuronal precursors.
Differential usage of DNA repair pathways at different developmental stages, associated with a different apoptotic threshold, may justify most of the specific effects of GIN. For instance, mice defective for NHEJ because of DNA ligase IV (Lig4) inactivation show DNA-damage-induced apoptosis in post-mitotic differentiating immature neurons, while HRR-deficient Xrcc2 knockout mice show apoptosis only in neural precursors cells, thus suggesting that during early development, DSB are repaired mainly using HRR [27]. Another example of differential requirement of DNA-damage response (DDR) factors during development is the conditional ablation of Topbp1, which is an activator of ATR during DDR and an important factor to maintain DNA integrity during replication. Deletion of Topbp1 in early cortical progenitors, using an Emx1-Cre driver, leads to microcephaly and strong apoptosis during development. In contrast, deletion at later stages using Nes-Cre results in similar levels of DNA damage, but attenuated apoptosis. In addition to the temporally restricted dependence on different DNA repair pathways, these data suggest that neural progenitors have different thresholds for apoptosis at different developmental stages and support the idea that apoptosis in early progenitors might be a crucial mechanism to ensure genome integrity in CNS [28, 29, 33].
Neurons display high sensitivity to GIN even after the end of development. The high levels of oxidative metabolism that characterize brain produce a high burden of reactive oxygen species, which are the main cause of SSB. Transcription-associated breaks [34] and neurotransmission-associated stress are additional SSB sources [35]. Accordingly, most patients bearing mutations in gene responsible to sense or repair SSB display neurological syndromes [33].
DNA-damage-sensing pathways and cortical development
In all tissues, DNA damage is sensed by genome surveillance pathways that may prevent cell-cycle progression, or initiate apoptosis, thus allowing DNA repair or preventing the potential danger posed by damaged cells [36]. Genome surveillance is ensured by three PI3K-like protein kinases: ATM (ataxia–telangiectasia, mutated), ATR (ATM and Rad3-related), and PRKDC (the catalytic subunit of the DNA-dependent protein kinase) [23]. The relative importance of individual kinases in CNS development is dependent on the type of DNA lesion and cells [37]. ATR exerts a prominent role in protecting cells from DNA damage arising during S phase in consequence of DNA stressors like ultraviolet radiation, DNA polymerase inhibitors, nucleotide depletion, or DNA crosslinkers, leading to DNA polymerase slowing or stalling [23]. These agents converge on formation of single-stranded DNA (ssDNA) stretches at the replication fork, which may produce replication intermediates leading to high risks of mutation and/or genomic rearrangements. Canonical ATR pathway activation is triggered when the replication protein A complex (RPA) binds to ssDNA, generating a recruitment platform for different proteins, including ATR-interacting protein (ATRIP), which in turn recruits ATR. In addition to ATR–ATRIP complex formation, full ATR activation requires the simultaneous presence on ssDNA of the heterotrimeric ring-shaped complex RAD9–RAD1–HUS1 [38]. This complex locally recruits TOPBP1 that binds to and activates ATR, leading to phosphorylation of checkpoint kinase-1 (CHK1) and other downstream ATR effectors, which slow origin firing, arrest cell cycle, stabilize, and restart stalled replication forks [39]. In contrast, ATM and PRKDC are activated by DNA double-strand breaks (DSBs) and exert a pivotal function in DSB recognition [23]. When DSB are recognized by the MRN complex (MRE11–RAD50–NBS1), ATM is recruited at DNA-damage sites, leading to phosphorylation of H2AX histone variant at S139 (γH2Ax) in proximity of the break. γH2Ax binds to MDC1, which further amplifies the signal by recruiting more MRN molecules and TP53BP1 [40]. ATM activation is crucial in preventing DNA-damaged cells from entering S phase. ATM also activates TP53, which in turn induces cell-cycle blocker CDKN1A (p21CIP1) as well as apoptosis-initiating genes. Most DSB occurring outside S phase are repaired by NHEJ independently of ATM, but a minority of DSBs located in heterochromatic regions or with blocked ends require processing in an ATM-dependent fashion. PRKDC is engaged to DSBs by the heterodimer complex KU70/80 to form the DNA-PK holoenzyme and facilitate DNA repair via NHEJ [41]. However, NHEJ core components’ inactivation leads to extensive neural apoptosis and embryonic lethality, while mice-lacking PRKDC are viable [42, 43]. How these three kinases collectively organize the responses to DNA damage has been elegantly demonstrated by Enriquez-Rios and colleagues: mice-bearing single or combined mutations of Atm, Atr, or Prkdc showed differential cellular sensitivity in response to IR [37]. Authors confirmed that ATM transduces pro-apoptotic signals in non-cycling cells, while it turned out to be less important in replicating progenitors in VZ. In contrast, ATR deletion did not affect the increased apoptosis induced by IR in the presence of PRKDC loss. Interestingly, apoptosis levels were increased by PRKDC loss even when both ATM and ATR were deleted (triple knockout mice), implying the existence of an additional, non-defined pathway converging on TP53 activation. In humans, PRKDC mutations have been associated to a rare syndrome characterized by immune deficiency and microcephaly [44, 45]. ATM loss is responsible for the neurodegenerative syndrome ataxia–telangiectasia, while hypomorphic ATR mutations are associated with one of the complementation groups of Seckel syndrome (SCKS) [29, 46] (see below).
In summary, the DNA-damage sensors ATR and PRKDC appear to be crucial for coping with endogenous and exogenous replication-related stress arising during embryonic development. Neural tissue could be affected more than other districts because of the stress imposed by the explosive proliferation of neural progenitors that especially characterize late-born neurons [47]. On the other hand, ATM is much more important for post-mitotic neurons, but the reasons for this specificity remain undefined.
DNA repair pathways in cortical development
In human gestation, cortical neurogenesis lasts for a long period, spanning from 7th until 28th week [48]. Neural progenitors, committed progenitors and differentiated cells are continually under threat by lesions from endogenous or exogenous sources. Base modifications, similar to those induced by ultraviolet radiation, may distort the helical structure of the DNA and are repaired by the nucleotide excision repair pathway (NER), while DNA base modifications caused by alkylation, deamination, or oxidation, such as those induced by reactive oxygen species (ROS), are mainly repaired by the base excision repair pathway (BER). DNA double-strand breaks (DSB) are the most harmful type of DNA lesion, which may lead to apoptosis or mutagenic rearrangements, such as translocations if left unrepaired or not correctly repaired [36]. DSBs may be generated exogenously, by Ionizing Radiations (IR) or different chemical agents (such as chemotherapeutic drugs), or endogenously, by prolonged replication fork stalling or ROS [49]. DSBs can be managed by cells through two different core pathways: HRR and NHEJ [50].
Base excision repair
The most common DNA lesion, DNA single-strand break (SSB), may be produced by ROS reaction with the deoxyribose sugar of DNA, as normal enzymatic intermediate of Base Excision Repair (BER) pathway, from catalytic intermediates generated by topoisomerase 1 (TOP1) to resolve supercoiled structures during replication, transcription, or other events. Given the different sources of SSBs, chemical composition of SSB ends can be highly heterogeneous and a wide variety of sub-pathways of SSB repair have evolved. Persistent SSBs may lead to replication fork collapse during chromosome duplication with consequent formation of DSBs, as well as transcription block in non-proliferating cells [33]. To deal with oxidation, deamination and spontaneous hydrolysis of bases, specific subtypes of BER operate during all stages of the cell cycle and exert a key function in both dividing and non-dividing cells. The sequence of BER is started by a lesion-specific DNA glycosylase, which recognizes and hydrolyzes the N–glycosidic bond of a substrate base, generating an apurinic/apyrimidinic site intermediate. This alterations, which can be frequently formed by spontaneous or damage-induced hydrolysis of the N–glycosidic bond, are cut by an endonuclease, APEX1. This protein works in coordination with PNKP to generate 5′-P and 3′-OH priming group, for repair synthesis and ligation. BER then engages Pol β, to replace the missing nucleotide (short patch repair) or Pol δ/ε in collaboration with PCNA, to perform displacing synthesis during S phase or when ATP concentrations are low (long patch repair). Finally, the repair factor complex XRCC1 and XRCC1 and DNA ligases LIG1 or LIG3 seal the remaining nick [51]. Mutations in genes encoding for BER components such as APTX, TDP1, or PNKP may produce different CNS phenotypes. TDP1 mutations cause a neurodegenerative disorder, Spinocerebellar ataxia with axonal neuropathy (SCAN1) [52], while mutations in APTX are responsible for Ataxia–ocular motor apraxia 1 (AOA1) [53]. PNKP mutations may result in neurodegeneration (AOA4) [54], but may also cause a neurodevelopmental disorder, Microcephaly with Seizures (MCSZ) [55]. This heterogeneity could be explained by the dual role played by PNKP, which is a key factor in BER, but is also involved in NHEJ through its interaction with XRCC4. PNKP is thus involved in repair of both SSBR and DSBR [33, 56], but it is likely to play a role only in a fraction of breaks repaired by NHEJ after IR damage [57].
Nucleotide excision repair
Nucleotide excision repair (NER) is a repair system capable of removing a wide variety of DNA helix-distorting lesions, such as base modifications or UV-induced photolesions and DNA adducts induced by chemicals like N-acetoxy-2-acetylaminofluorene or aflatoxin B1. Patients bearing congenital defects in NER components are peculiarly sensitive to sunlight, and suffer for cancer predisposition and/or premature aging [58].
NER pathway is very important during neural progenitor proliferation, as NER can cope with stalled RNA polymerase or abortive transcripts, monitor DNA integrity, and activate DNA-damage signaling. In this repair system, a central role is exerted by components of the basal TFIIH transcription and repair complex, required for RNA pol II function. This system is responsible for transcription-coupled repair (TCR), as well as for transcription-independent global genomic repair (GGR). It is worth noticing that mutations involving TCR lead to neuropathology, while those affecting GGR have much milder neurological implications. The two pathways share a high number of repair factors but differ during the DNA-damage recognition phase [49]. In TCR, DNA lesions affect a transcribed gene and are sensed in consequence of RNA Pol II stalling. In this case, the repair process is initiated by ERCC6 that, in turn, recruits ERCC8. Instead, the proteins required to recognize DNA damage in GGR are XPC and XPE. For both pathways, DNA-damage verification and unwinding is carried out by TFIIH, a DNA-dependent ATPase and helicase activity that, in complex with ERCC3 or ERCC2, opens up the DNA helix around the lesion. Finally, the DNA strand that contain the lesion is cut at the single-to-double-strand DNA transitions by the structure-specific endonucleases ERCC5 and ERCC1, gap filling is carried out by DNA Pol δ/ε using undamaged strand as a template, and the nick is sealed by DNA ligase I or III [59]. Mutations in several components of the NER pathway can lead to the human syndromes Xeroderma Pigmentosum (XP), Cockayne Syndrome (CS), and Trichothiodystrophy (TTD). Defects in NER components may lead to a complex relationship between genetic alterations and clinical phenotype [60]. While mutations in different genes, grouped in eight XP complementation groups, lead to very similar phenotypes, different mutations in just one gene (ERCC2) may lead to six different clinical disorders: XP, XP with neurological disease, TTD, XP/CS complex, XP/TTD complex, or cerebro-oculo-facio-skeletal syndrome. XP is rare disease affecting the skin and leading to neurodegeneration (that results in brain atrophy). Exposure of these patients to sunlight results in a high incidence of skin and mucous membrane cancer, squamous and basal cell carcinomas and melanomas [61]. However, mutations in ERCC8 or ERCC6, lead to CS. CS is a developmental and neurological disorder, associated with reduced lifespan without an increased incidence of cancer. In this case, neuropathology is profoundly different from that associated with XP, as CS is characterized by microcephaly with intellectual disability. Microcephaly in CS is likely due to combination of defective cortical development and progressive brain atrophy. In addition, vasculopathy and calcification of the basal ganglia may also be observed. To complicate even further the situation, CS may be caused by mutations in ERCC2, ERCC3, or ERCC5 [62]. TTD is a rare autosomal recessive disorder characterized by sulphur deficient brittle hair, sometimes accompanied by facial dysmorphism and intellectual disability. TTD arises from mutations in the gene-encoding ERCC2 helicase or, less frequently, ERCC3 or GTF2H5 proteins. The neurological abnormalities observed in TTD include developmental microcephaly associated with intellectual disability, deafness, and ataxia [63].
Similar to CS, TTD may further affect cortical development by altering myelin formation. Moreover, neurological defects observed in TTD patients may be due to the deregulation of thyroid hormone target genes in the brain [64].
Homologous recombination repair and Fanconi anemia pathways
In cycling neural progenitors, genomic fidelity during replication is ensured by HRR. HRR is considered an error-free mechanism, occurring during S or G2 phase of the cell cycle. During the initial stages of HRR, ATM and ATR kinases recognize double-strand DNA break and phosphorylate downstream targets such as H2AX, BRCA1, CHEK2, and TP53. BRCA1, along with BARD1 and BRIP1, acts as a scaffold that coordinates proteins at the damage site [65]. HRR requires a sister chromatid as a template to repair DNA and is initiated by the MRE11-RAD50-NBS1 complex, in collaboration with RBBP8, which generates 3′ ssDNA overhang for strand exchange [66]. MRN complex participates in both HR and NHEJ (see below); in contrast RBBP facilitates HR while suppressing NHEJ [67] [68]. RAD51 is recruited to 3′ ssDNA overhangs through a complex mechanism involving the RPA, BRCA2, BRCA1, and XRCC2 proteins, to form RAD51 nucleoprotein filaments, which are essential for strand invasion of the intact homologous DNA region used as a template for repair. Studies in knockout mouse models have shown that HRR genes are fundamental for brain development [69–71]. In humans, mutations in these genes have been associated with heterogeneous clinical manifestations, which may (BRCA1) [72, 73] or may not (BRCA2) include microcephaly [74, 75]. One of the peculiar phenotypes that may derive from HRR pathway mutations is Fanconi anemia (FA), a clinically and genetically heterogeneous disorder mainly characterized by bone-marrow failure and cancer predisposition. However, some patients also display microcephaly and dwarfism features, reminiscent of Seckel syndrome [76]. To date, at least eighteen FANC genes are associated with patient mutations. The FA nuclear complex, composed of FANCA, FANCC, FANCG, and FANCF proteins, is essential for protection against chromosome breakage. The core complex of the FA pathway is necessary to monoubiquitylate a specific site in each protein of the FANCI/FANCD2 (ID2) heterodimer. Monoubiquitination of ID2 triggers nucleases SLX4 and ERCC4, as well as downstream repair factors that include BRIP, BRCA2, PALB2, and RAD51C [77]. The latter two genes are in common with the HRR pathway. FA proteins have a prominent role in promoting the repair of DNA inter-strand crosslinks (ICLs) [78]. Unrepaired ICLs are very harmful, especially in dividing cells, as they prevent strand separation, thereby stalling DNA replication and leading to chromosomal instability [79]. FA pathway is closely dependent on ATR and CHEK1 function. Indeed, ATR deficient cells are hypersensitive to ICLs, and many FA proteins, such as FANCD2, FANCA, FANCI, and FANCE, are direct substrates of ATR or CHEK1 [80].
Non-homologous end joining
NHEJ, which works in both non-replicating and replicating cells, is the predominant DSB repair system in higher eukaryotes. NHEJ involves ligation of two broken DNA ends without needing a repair template. NHEJ core factors, including XRCC4, LIG4, and NHEJ1, associated with DSB after PRKDC recruits Ku proteins to the broken ends, which are subsequently closely aligned and ligated. The repair of some DSBs by NHEJ may require additional accessory proteins, such as DNA polymerases and nucleases. One of such factors is DCLRE1C, an endonuclease interacting with PRKDC to promote repair of a subset of DSBs [81–83]. Indeed, PRKDC recruits DCLRE1C to promote DNA-end processing [84, 85].
Although NHEJ is often described as an error prone and mutagenic process, it is more efficient and accurate than previously thought [86]. Only when classical NHEJ (cNHEJ) fails, cells use an alternative end-joining pathway mediated by DNA polymerase theta, the so-called theta-mediated end-joining (TMEJ), which can easily introduce mutations [87, 88]. The study of mice mutated for genes important in NHEJ [89] showed elevated apoptosis and enhanced IR sensitivity in cortical progenitors. NHEJ is the main repair pathway in differentiating and post-mitotic neurons [27]. NHEJ is also required during immune system development for V(D)J and Class Switch Recombination mechanisms [90]. For these reasons, congenital defects in NHEJ pathway components such PRKDC [45], LIG4 [91] and gene-encoding XRCC4 and NHEJ1 [75, 92] are associated with developmental delay, malignancy, variable degree of immunodeficiency and microcephaly.
The MCPH–SCKS spectrum: many roads leading from genomic instability to primary microcephaly
Severe congenital microcephalies are characterized by reduced head circumference at birth, to at least three standard deviations below the mean [1, 93]. The two main clinical phenotypes are referred to as primary hereditary microcephaly (MCPH) and Seckel syndrome (SCKS) [1, 93]. In MCPH, brain size reduction is the main clinical feature, associated with conserved brain architecture and mild-to-moderate intellectual disability. MCPH is a genetically heterogeneous condition, with at least 18 complementation groups. Most of the genetic defects identified in MCPH involve genes that play fundamental roles in various processes that collectively enable cells to faithfully segregate chromosomes and allow correct progression of mitotic division [94]. These defects may range from abnormal microtubule formation, stabilization, and polymerization, to alteration of spindle structure, defective centrosome function, along with impaired cell-cycle checkpoint function. Although these disfunctions may all indirectly lead to CIN and GIN (see below), some of MCPH genes have a more direct involvement in DNA repair and chromosome stability. Under this point of view, the case of MCPH1 is particularly striking, because this multifunctional gene may affect diverse aspects of cell-cycle regulation and integrity. MCPH1 [95, 96] encodes for a centrosomal protein containing three BRCT (BRCA1 C-terminal) domains. Various studies demonstrated that MCPH1 plays an important role in G1/S and G2/M checkpoint regulation and in maintaining telomere integrity [97]. MCPH1-deficient cells exhibit mitotic defects, such as a prophase-like state and premature chromosome condensation [98] [99]. MCPH1 plays a crucial role in different aspects of DNA-damage response downstream of both ATM and ATR pathways. Indeed, MCPH1 is concentrated at DNA-damage foci in response to UV or IR treatment, colocalizing with γH2AX, TP53BP1, pATM, ATR, and RPA. In MCPH1-deficient cells, the IR-induced formation of TP53BP1 and pATM foci or RPA recruitment to ssDNA is impaired. ATR signaling amplification under replication stress requires MCPH1 to recruit TOPBP1 [100]. Moreover, during HRR, MCPH1 binds to BRCA2 and regulates the localization of BRCA2 and RAD51 at DNA-damage sites [101]. In addition, MCPH1 transcriptionally regulates CHK1 and BRCA1 expression, by interacting with E2F1 on their promoters [102]. Finally, MCPH1 directly acts on chromatin structure, facilitating the access of repair proteins to DNA-damage sites, by interacting with the chromatin-remodeling complex SWI/SNF in an ATR-dependent manner [103]. Chromatin remodeling is also regulated by PHC1, a human homologue of the Drosophila Polyhomeotic gene, which takes part to the Polycomb Repressive Complex 1 and is mutated in MCPH11. Cells defective for PHC1 show enhanced levels of the DNA replication inhibitor GMNN, accumulate DNA damage and are defective in DNA repair and proliferation after IR treatment [104]. A direct involvement in GIN has recently been proposed for ASPM, the most frequently mutated MCPH gene [105]. ASPM-dependent microcephaly (MCPH5) is commonly believed to result from abnormal spindle function and positioning, leading to the alteration of symmetric to asymmetric cell division [106]. However, ASPM levels are influenced by IR and ASPM affects efficient DSB repair and reduces DNA damage during cerebellar granule neuron progenitors cell proliferation [107] [108]. Another multifunctional gene causing MCPH is Citron Rho-Interacting Kinase (CIT), encoding Citron kinase protein (CITK). CITK localizes at the centrosomes and midbody, where it plays a major role in regulating the stability of the microtubule cytoskeleton. Mutation of CIT leads to apoptosis in neural progenitors, newborn neurons and in male germ cells [109–111], which was initially proposed to result from cytokinesis failure [112]. In addition, mouse Cit-mutant neural progenitors show aberrant cleavage plane orientation [113] and accumulate DNA double-strand beaks [114]. The latter phenotype correlates with defective RAD51 localization to DNA-damage foci and is probably responsible of the apoptotic phenotype through TP53 activation [114]. Altogether, these studies underscore that maintenance of genome stability is a critical functional aspect common to many of MCPH genes.
On the other hand, SCKS is characterized by intrauterine growth retardation, severe proportionate short stature, profound microcephaly with intellectual disability and characteristic craniofacial features, in the absence of visceral malformations.
In addition to ATR mutation, SCKS can be caused by mutation of RBBP8, CENPJ, CEP152, CEP63, NIN, DNA2, TRAIP, and NSMCE2. However, patients carrying mutations in some of these genes may show phenotypes intermediate between SCKS and MCPH, or even pure MCPH. These genes include RBBP8, CEP152 (involved in MCPH9), CENPJ (involved in MCPH6), CEP63, and PHC1 (involved in MCPH11). Therefore, while in the past SCKS and MCPH were discriminated by height, the most recent view is that the two phenotypes define the extremes of a spectrum of phenotypes in which genomic stability of neural progenitors is mostly compromised.
Centrosomes and chromosome segregation at the core of genome stability and neurologic disease
Centrosome is the main microtubule-organizing center (MTOC), essential for coordinating cell-cycle progression, mitotic cell division, cilia formation, and DNA-damage response [115]. Moreover, in cortical neural progenitors, centrosome plays a pivotal role in regulating cleavage plane orientation [116], cell fate decision, and neuronal migration [117]. During mitosis, an intact centrosome number is pivotal for correct chromosome segregation and to maintain genomic integrity. Alteration of centrosome number and function can impact on genome stability in different ways. Increased centrosome number may lead to multipolar mitoses, which may result in mitotic catastrophe or undergo bipolar resolution [118]. Even in this case, there is a high frequency of abnormal microtubule to kinetochore attachments, resulting in chromosome mis-segregation [118]. The latter event may lead to aneuploidy, DNA damage through micronuclei formation, or even cytokinesis failure ending up with polyploidy [119]. These defects can result from either complete loss of centrosomes, disruption of normal centrosome activity, or from defects in centrosome separation [120]. All these events may have a profound importance on cortical development [121–123].
Therefore, it is not surprising that mutations in genes encoding structural centrosome components or centrosome-associated proteins are the most frequent cause of MCPH (CEP152, CEP63, SASS6, STIL, CENPJ, CEP135, CDK5RAP2, CDK6, ASPM, and WDR62), and Microcephalic primordial dwarfism (MPD) [1, 124, 125]. The importance of correct chromosome segregation is further suggested by the occurrence of microcephaly in the so-called “condensinopathies”, i.e., genetic syndromes deriving from mutations in components of condensin complexes (NCAPD2, NCAPH or NCAPD3) [126]. Condensinopathies are characterized by inappropriate chromosome compaction and decatenation, chromosome segregation errors and micronucleus formation, with consequent increased frequency of aneuploidy [126]. Of note, the function of Condensin II complex is regulated by MCPH1, explaining the premature chromosome condensation (PCC) defect observed in cells derived from MCPH1 patients [127]. It is also emerging that centrosomes could be important integration points for DNA repair pathways and control cell-cycle progression after DNA damage. Indeed, some DDR players such as ATM, ATR, CHEK2, CHEK1, BRCA1, BRCA2 and PARPs may localize to the centrosome. Centrosomal localization of CHEK1 is important to inactivate the phosphatase CDC25, which is an activator of CDK1-cyclinB, the M-phase promoting factor [128]. MCPH1 controls the localization of CHEK1 to centrosomes and its loss causes premature CHEK1 activation and early mitotic entry, thus uncoupling mitosis and the centrosome cycle [129]. Moreover, TP53, a common effector of the DNA-damage response implicated in cell-cycle progression, DNA repair and apoptosis, can localize to centrosome after DNA damage, in an ATM-dependent manner [130]. Another important link between DDR and centrosome is the observation that nuclear-cytoplasmic trafficking of DDR proteins occurs on microtubules and is dependent on dynein. Interestingly, microtubule-targeting agents (e.g., taxanes), can alter the intracellular distribution of several DDR proteins (ATM, ATR, PRKDC, MRE11, RAD50, TP53, NBS1, TP53BP1, and TP63) [131]. Finally, microtubules are required to control chromatin mobility, in particular in case of DNA break, where an enhanced chromatin moving seems to be important to “expose” the damage and allow DDR factors to reach and repair the lesion [132]. An additional link between centrosome and DNA-damage response is represented by SCKS and FA, which display centrosomal disfunction and DDR impairment. SCKS may arise from mutations in centrosomal proteins CEP152 or PLK4 [133–135], leading to impaired centrosome biogenesis, supernumerary centrosomes and impaired cell-cycle progression, together with genomic instability and defective DDR. Loss of FA signaling can weaken the spindle assembly checkpoint (SAC), which in normal conditions prevents separation of the duplicated chromosomes until each chromosome is properly attached to the spindle apparatus. FA deficient cells display chromosome instability caused by abnormal cell-cycle progression, cytokinesis failure and accumulation of ultrafine bridges. Moreover, FA deficient cells accumulate DNA damage, since they are hypersensitive to some metabolic by-products such as aldehydes [76].
Perspectives and conclusions
Knowledge of the fundamental processes underlying brain cortex development has grown exponentially during the last few years. These studies have underscored the crucial relevance for brain development and physiology of the different pathways that have evolved to ensure faithful duplication, transmission and maintenance of the genomic information. At the same time, these studies have started to unravel the complex interrelation between the molecular details of these pathways and the phenotypic spectrum of CNS abnormalities produced by their alterations. Of particular relevance, the centrosome is progressively emerging as a possible integration hub for molecular events specifically important for neurogenesis. It is, therefore, easy to foresee that the study of genome stability maintenance in CNS will be a very interesting topic for future studies. Among the most interesting challenges, it will be important to determine how centrosome functions and DDR are finely orchestrated and why neurogenesis is so sensitive to their alterations. Moreover, it appears very important to better define the role of microtubules in this context. A deeper knowledge of these phenomena could be critical to better understand why alterations of apparently different pathways produce similar phenotypes, as well as how different mutations of the same gene may result in completely different pathologies.
Acknowledgements
We apologize with Colleagues whose work could not be mentioned due to space constraints. The financial support to F.D.C. laboratory by Associazione Italiana per la Ricerca sul Cancro (AIRC—Grant IG17527), by Telethon Foundation (Grant nos. GGP12095 and GGP13081), by ‘Consiglio Nazionale delle Ricerche’ through the EPIGEN project and by the Jérôme Lejeune Foundation is gratefully acknowledged.
References
- 1.Passemard S, Kaindl AM, Verloes A. Microcephaly. Handb Clin Neurol. 2013;111:129–141. doi: 10.1016/B978-0-444-52891-9.00013-0. [DOI] [PubMed] [Google Scholar]
- 2.Cheeran MC-J, Lokensgard JR, Schleiss MR. Neuropathogenesis of congenital cytomegalovirus infection: disease mechanisms and prospects for intervention. Clin Microbiol Rev. 2009;22:99–126. doi: 10.1128/cmr.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duerinckx S, Abramowicz M. The genetics of congenitally small brains. Semin Cell Dev Biol. 2018;76:76–85. doi: 10.1016/j.semcdb.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Del Campo M, Jones KL. A review of the physical features of the fetal alcohol spectrum disorders. Eur J Med Genet. 2017;60:55–64. doi: 10.1016/j.ejmg.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial–cell specification. Nature. 2010;468:214. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 6.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-P. [DOI] [PubMed] [Google Scholar]
- 7.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 8.Campbell K, Götz M. Radial glia: multi-purpose cells for vertebrate brain development. Trends Neurosci. 2002;25:235–238. doi: 10.1016/S0166-2236(02)02156-2. [DOI] [PubMed] [Google Scholar]
- 9.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Borrell V, Götz M. Role of radial glial cells in cerebral cortex folding. Curr Opin Neurobiol. 2014;27:39–46. doi: 10.1016/j.conb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 13.Aguilera A, García-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 14.Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol. 2013;425:4733–4744. doi: 10.1016/j.jmb.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchová Z. The presence of extra chromosomes leads to genomic instability. Nat Commun. 2016;7:10754. doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Andriani GA, Vijg J, Montagna C. Mechanisms and consequences of aneuploidy and chromosome instability in the aging brain. Mech Ageing Dev. 2017;161:19–36. doi: 10.1016/j.mad.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verreet T, Verslegers M, Quintens R, Baatout S, Benotmane MA. Current evidence for developmental, structural, and functional brain defects following prenatal radiation exposure. Neural Plast. 2016 doi: 10.1155/2016/1243527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood JW, Johnson KG, Omori Y, Kawamoto S, Keehn RJ. Mental retardation in children exposed in utero to the atomic bombs in Hiroshima and Nagasaki. Am J Public Health Nations Health. 1967;57:1381–1389. doi: 10.2105/AJPH.57.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci. 2009;10:100–112. doi: 10.1038/nrn2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowak E, Etienne O, Millet P, Lages CS, Mathieu C, Mouthon M-A, Boussin FD. Radiation-induced H2AX phosphorylation and neural precursor apoptosis in the developing brain of mice. Radiat Res. 2006;165:155–164. doi: 10.1667/RR3496.1. [DOI] [PubMed] [Google Scholar]
- 23.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 25.Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, Huttner WB. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair. 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci USA. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Katyal S, Downing SM, Zhao J, Russell HR, McKinnon PJ. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nat Neurosci. 2012;15:819–826. doi: 10.1038/nn.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Shull ERP, Frappart P-O, Katyal S, Enriquez-Rios V, Zhao J, Russell HR, Brown EJ, McKinnon PJ. ATR maintains select progenitors during nervous system development. EMBO J. 2012;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murga M, Bunting S, Montaña MF, Soria R, Mulero F, Cañamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe N, Mii S, Asai N, Asai M, Niimi K, Ushida K, Kato T, Enomoto A, Ishii H, Takahashi M, Murakumo Y. The REV7 subunit of DNA polymerase ζ is essential for primordial germ cell maintenance in the mouse. J Biol Chem. 2013;288:10459–10471. doi: 10.1074/jbc.M112.421966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:nrg2380. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 34.Marnef A, Cohen S, Legube G. Transcription-coupled DNA double-strand break repair: active genes need special care. J Mol Biol. 2017;429:1277–1288. doi: 10.1016/j.jmb.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Hetman M, Vashishta A, Rempala G. Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition. J Neurochem. 2010;114:1537–1549. doi: 10.1111/j.1471-4159.2010.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enriquez-Rios V, Dumitrache LC, Downing SM, Li Y, Brown EJ, Russell HR, McKinnon PJ. DNA-PKcs, ATM, and ATR interplay maintains genome integrity during neurogenesis. J Neurosci. 2017;37:893–905. doi: 10.1523/JNEUROSCI.4213-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Shell SM, Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene. 2005;24:4728–4735. doi: 10.1038/sj.onc.1208674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18:622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res Rev Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Jette N, Lees-Miller SP. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt FW. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 44.van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari P-O, Tezcan I, Chen DJ, Zdzienicka MZ, van Dongen JJM, van Gent DC. A DNA-PKcs mutation in a radiosensitive T-B-SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodbine L, Neal JA, Sasi N-K, Shimada M, Deem K, Coleman H, Dobyns WB, Ogi T, Meek K, Davies EG, Jeggo PA. PRKDC mutations in a SCID patient with profound neurological abnormalities. J Clin Invest. 2013;123:2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair. 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 47.Swahari V, Nakamura A, Baran-Gale J, Garcia I, Crowther AJ, Sons R, Gershon TR, Hammond S, Sethupathy P, Deshmukh M. Essential function of dicer in resolving DNA damage in the rapidly dividing cells of the developing and malignant cerebellum. Cell Rep. 2016;14:216–224. doi: 10.1016/j.celrep.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyama T, Wilson DM. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Driscoll M, Jeggo PA. The role of double-strand break repair—insights from human genetics. Nat Rev Genet. 2006;7:nrg1746. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 51.Kim YJ, Wilson DM 3rd. Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5(1):3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MAM, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 53.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonça P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 54.Bras J, Alonso I, Barbot C, Costa MM, Darwent L, Orme T, Sequeiros J, Hardy J, Coutinho P, Guerreiro R. Mutations in PNKP cause recessive ataxia with oculomotor apraxia type 4. Am J Hum Genet. 2015;96:474–479. doi: 10.1016/j.ajhg.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, Ganesh VS, Chang BS, Grix A, Hill RS, Topcu M, Caldecott KW, Barkovich AJ, Walsh CA. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimada M, Dumitrache LC, Russell HR, McKinnon PJ. Polynucleotide kinase–phosphatase enables neurogenesis via multiple DNA repair pathways to maintain genome stability. EMBO J. 2015;34:2465–2480. doi: 10.15252/embj.201591363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edifizi D, Schumacher B. Genome instability in development and aging: insights from nucleotide excision repair in humans, mice, and worms. Biomolecules. 2015;5:1855–1869. doi: 10.3390/biom5031855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dijk M, Typas D, Mullenders L, Pines A. Insight in the multilevel regulation of NER. Exp Cell Res. 2014;329:116–123. doi: 10.1016/j.yexcr.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and cockayne syndrome: a complex genotype–phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam ET, Revet I, Cleaver JE. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 62.Rapin I. Disorders of nucleotide excision repair. Handb Clin Neurol. 2013;113:1637–1650. doi: 10.1016/B978-0-444-59565-2.00032-0. [DOI] [PubMed] [Google Scholar]
- 63.Faghri S, Tamura D, Kraemer KH, DiGiovanna JJ. Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J Med Genet. 2008;45:609–621. doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borrelli E, Compe E, Marescaux J, Egly J-M, Soler L, Malerba M. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat Neurosci. 2007;10:1414. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]
- 65.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 66.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:nature06337. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JTF, Tkáč J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 68.Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol Cell. 2016;64:940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 69.Pao GM, Zhu Q, Perez-Garcia CG, Chou S-J, Suh H, Gage FH, O’Leary DDM, Verma IM. Role of BRCA1 in brain development. Proc Natl Acad Sci USA. 2014;111:E1240–E1248. doi: 10.1073/pnas.1400783111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frappart P-O, Lee Y, Lamont J, McKinnon PJ. BRCA2 is required for neurogenesis and suppression of medulloblastoma. EMBO J. 2007;26:2732–2742. doi: 10.1038/sj.emboj.7601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, Monteiro ANA, Messick TE, Powers J, Yonker A, Couch FJ, Goldgar DE, Davidson HR, Nathanson KL, Foulkes WD, Greenberg RA. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 2013;3:399–405. doi: 10.1158/2159-8290.CD-12-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freire BL, Homma TK, Funari MFA, Lerario AM, Leal AM, Velloso EDRP, Malaquias AC, Jorge AAL. Homozygous loss of function BRCA1 variant causing a Fanconi-anemia-like phenotype, a clinical report and review of previous patients. Eur J Med Genet. 2017 doi: 10.1016/j.ejmg.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Rump P, Jazayeri O, van Dijk-Bos KK, Johansson LF, van Essen AJ, Verheij JBGM, Veenstra-Knol HE, Redeker EJW, Mannens MMAM, Swertz MA, Alizadeh BZ, van Ravenswaaij-Arts CMA, Sinke RJ, Sikkema-Raddatz B. Whole-exome sequencing is a powerful approach for establishing the etiological diagnosis in patients with intellectual disability and microcephaly. BMC Med Genom. 2016;9:7. doi: 10.1186/s12920-016-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaheen R, Faqeih E, Ansari S, Abdel-Salam G, Al-Hassnan ZN, Al-Shidi T, Alomar R, Sogaty S, Alkuraya FS. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res. 2014;24:291–299. doi: 10.1101/gr.160572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nalepa G, Clapp DW. Fanconi anemia and the cell cycle: new perspectives on aneuploidy. F1000prime Rep. 2014;6:23. doi: 10.12703/p6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhattacharjee S, Nandi S. DNA damage response and cancer therapeutics through the lens of the Fanconi anemia DNA repair pathway. Cell Commun Signal CCS. 2017;15:41. doi: 10.1186/s12964-017-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hashimoto S, Anai H, Hanada K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016 doi: 10.1186/s41021-016-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:nrm.2016.48. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 80.Kee Y, D’Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rooney S, Alt FW, Lombard D, Whitlow S, Eckersdorff M, Fleming J, Fugmann S, Ferguson DO, Schatz DG, Sekiguchi J. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J Exp Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 83.Riballo E, Kühne M, Rief N, Doherty A, Smith GCM, Recio M-J, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Löbrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 84.Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, Lee BJ, Dubois RL, Liu C, Yu X, Lan L, Zha S. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol Cell. 2015;58:172–185. doi: 10.1016/j.molcel.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malu S, De Ioannes P, Kozlov M, Greene M, Francis D, Hanna M, Pena J, Escalante CR, Kurosawa A, Erdjument-Bromage H, Tempst P, Adachi N, Vezzoni P, Villa A, Aggarwal AK, Cortes P. Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA ligase IV and DNA-PKcs. J Exp Med. 2012;209:955–963. doi: 10.1084/jem.20111437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bétermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Schendel R, Roerink SF, Portegijs V, van den Heuvel S, Tijsterman M. Polymerase Θ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat Commun. 2015;6:7394. doi: 10.1038/ncomms8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schimmel J, Kool H, van Schendel R, Tijsterman M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017 doi: 10.15252/embj.201796948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gatz SA, Ju L, Gruber R, Hoffmann E, Carr AM, Wang Z-Q, Liu C, Jeggo PA. Requirement for DNA ligase IV during embryonic neuronal development. J Neurosci. 2011;31:10088–10100. doi: 10.1523/JNEUROSCI.1324-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 91.Altmann T, Gennery AR. DNA ligase IV syndrome; a review. Orphanet J Rare Dis. 2016 doi: 10.1186/s13023-016-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche M-C, Sanal O, Plebani A, Stéphan J-L, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay J-P, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 93.Verloes A, Drunat S, Gressens P, Passemard S. Primary autosomal recessive microcephalies and Seckel syndrome spectrum disorders. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Mefford HC, Stephens K, Amemiya A, Ledbetter N, editors. GeneReviews(®) Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 94.Alcantara D, O’Driscoll M. Congenital microcephaly. Am J Med Genet C Semin Med Genet. 2014;166C:124–139. doi: 10.1002/ajmg.c.31397. [DOI] [PubMed] [Google Scholar]
- 95.Jackson AP, McHale DP, Campbell DA, Jafri H, Rashid Y, Mannan J, Karbani G, Corry P, Levene MI, Mueller RF, Markham AF, Lench NJ, Woods CG. Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet. 1998;63:541–546. doi: 10.1086/301966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, Karbani G, Jafri H, Rashid Y, Mueller RF, Markham AF, Woods CG. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venkatesh T, Suresh PS. Emerging roles of MCPH1: expedition from primary microcephaly to cancer. Eur J Cell Biol. 2014;93:98–105. doi: 10.1016/j.ejcb.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 98.Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, Neitzel H, Jackson AP. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–266. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neitzel H, Neumann LM, Schindler D, Wirges A, Tönnies H, Trimborn M, Krebsova A, Richter R, Sperling K. Premature chromosome condensation in humans associated with microcephaly and mental retardation: a novel autosomal recessive condition. Am J Hum Genet. 2002;70:1015–1022. doi: 10.1086/339518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X, Zhou Z-W, Wang Z-Q. The DNA damage response molecule MCPH1 in brain development and beyond. Acta Biochim Biophys Sin. 2016;48:678–685. doi: 10.1093/abbs/gmw048. [DOI] [PubMed] [Google Scholar]
- 101.Wu X, Mondal G, Wang X, Wu J, Yang L, Pankratz VS, Rowley M, Couch FJ. Microcephalin regulates BRCA2 and Rad51-associated DNA double-strand break repair. Cancer Res. 2009;69:5531–5536. doi: 10.1158/0008-5472.CAN-08-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang S-Z, Lin F-T, Lin W-C. MCPH1/BRIT1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep. 2008;9:907–915. doi: 10.1038/embor.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peng G, Yim E-K, Dai H, Jackson AP, van der Burgt I, Pan M-R, Hu R, Li K, Lin S-Y. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Awad S, Al-Dosari MS, Al-Yacoub N, Colak D, Salih MA, Alkuraya FS, Poizat C. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum Mol Genet. 2013;22:2200–2213. doi: 10.1093/hmg/ddt072. [DOI] [PubMed] [Google Scholar]
- 105.Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida GH, Hennekam RCM, Maher ER, Fryns J-P, Alswaid A, Jafri H, Rashid Y, Mubaidin A, Walsh CA, Roberts E, Woods CG. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kato TA, Okayasu R, Jeggo PA, Fujimori A. ASPM influences DNA double-strand break repair and represents a potential target for radiotherapy. Int J Radiat Biol. 2011;87:1189–1195. doi: 10.3109/09553002.2011.624152. [DOI] [PubMed] [Google Scholar]
- 108.Williams SE, Garcia I, Crowther AJ, Li S, Stewart A, Liu H, Lough KJ, O’Neill S, Veleta K, Oyarzabal EA, Merrill JR, Shih Y-YI, Gershon TR. Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Dev Camb Engl. 2015;142:3921–3932. doi: 10.1242/dev.124271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sgrò F, Bianchi FT, Falcone M, Pallavicini G, Gai M, Chiotto AMA, Berto GE, Turco E, Chang YJ, Huttner WB, Di Cunto F. Tissue-specific control of midbody microtubule stability by Citron kinase through modulation of TUBB3 phosphorylation. Cell Death Differ. 2016;23:801–813. doi: 10.1038/cdd.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cunto FD, Imarisio S, Camera P, Boitani C, Altruda F, Silengo L. Essential role of citron kinase in cytokinesis of spermatogenic precursors. J Cell Sci. 2002;115:4819–4826. doi: 10.1242/jcs.00163. [DOI] [PubMed] [Google Scholar]
- 111.Bianchi FT, Gai M, Berto GE, Di Cunto F. Of rings and spines: the multiple facets of Citron proteins in neural development. Small GTPases. 2017 doi: 10.1080/21541248.2017.1374325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A, Atzori C, Turco E, Triolo R, Dotto GP, Silengo L, Altruda F. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/S0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 113.Gai M, Bianchi FT, Vagnoni C, Vernì F, Bonaccorsi S, Pasquero S, Berto GE, Sgrò F, Chiotto AM, Annaratone L, Sapino A, Bergo A, Landsberger N, Bond J, Huttner WB, Di Cunto F. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep. 2016;17:1396–1409. doi: 10.15252/embr.201541823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bianchi FT, Tocco C, Pallavicini G, Liu Y, Vernì F, Merigliano C, Bonaccorsi S, El-Assawy N, Priano L, Gai M, Berto GE, Chiotto AMA, Sgrò F, Caramello A, Tasca L, Ala U, Neri F, Oliviero S, Mauro A, Geley S, Gatti M, Di Cunto F. Citron kinase deficiency leads to chromosomal instability and TP53-sensitive microcephaly. Cell Rep. 2017;18:1674–1686. doi: 10.1016/j.celrep.2017.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nano M, Basto R. Consequences of centrosome dysfunction during brain development. Adv Exp Med Biol. 2017;1002:19–45. doi: 10.1007/978-3-319-57127-0_2. [DOI] [PubMed] [Google Scholar]
- 116.Taverna E, Götz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- 117.Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011;48:349–358. doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 118.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hayashi MT, Karlseder J. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 2013;32:4593. doi: 10.1038/onc.2012.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lerit DA, Poulton JS. Centrosomes are multifunctional regulators of genome stability. Chromosome Res Int J Mol Supramol Evol Asp Chromosome Biol. 2016;24:5–17. doi: 10.1007/s10577-015-9506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lenroot RK, Lee NR, Giedd JN. Effects of sex chromosome aneuploidies on brain development: evidence from neuroimaging studies. Dev Disabil Res Rev. 2009;15:318–327. doi: 10.1002/ddrr.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dierssen M. Down syndrome: the brain in trisomic mode. Nat Rev Neurosci. 2012;13:844–858. doi: 10.1038/nrn3314. [DOI] [PubMed] [Google Scholar]
- 123.Rosenkrantz JL, Carbone L. Investigating somatic aneuploidy in the brain: why we need a new model. Chromosoma. 2017;126:337–350. doi: 10.1007/s00412-016-0615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zaqout S, Morris-Rosendahl D, Kaindl AM. Autosomal recessive primary microcephaly (MCPH): an update. Neuropediatrics. 2017;48:135–142. doi: 10.1055/s-0037-1601448. [DOI] [PubMed] [Google Scholar]
- 125.Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, Gerard B, Verloes A, Mani S, Gressens P. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 126.Martin C-A, Murray JE, Carroll P, Leitch A, Mackenzie KJ, Halachev M, Fetit AE, Keith C, Bicknell LS, Fluteau A, Gautier P, Hall EA, Joss S, Soares G, Silva J, Bober MB, Duker A, Wise CA, Quigley AJ, Phadke SR, Deciphering Developmental Disorders Study, Wood AJ, Vagnarelli P, Jackson AP Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016;30:2158–2172. doi: 10.1101/gad.286351.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yamashita D, Shintomi K, Ono T, Gavvovidis I, Schindler D, Neitzel H, Trimborn M, Hirano T. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol. 2011;194:841–854. doi: 10.1083/jcb.201106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23:3386–3396. doi: 10.1038/sj.emboj.7600328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart P-O, Wang Z-Q. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1–Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 130.Tritarelli A, Oricchio E, Ciciarello M, Mangiacasale R, Palena A, Lavia P, Soddu S, Cundari E. p53 localization at centrosomes during mitosis and postmitotic checkpoint are ATM-dependent and require serine 15 phosphorylation. Mol Biol Cell. 2004;15:3751–3757. doi: 10.1091/mbc.E03-12-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Poruchynsky MS, Komlodi-Pasztor E, Trostel S, Wilkerson J, Regairaz M, Pommier Y, Zhang X, Maity TK, Robey R, Burotto M, Sackett D, Guha U, Fojo AT. Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc Natl Acad Sci. 2015;112:1571–1576. doi: 10.1073/pnas.1416418112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lawrimore J, Barry TM, Barry RM, York AC, Friedman B, Cook DM, Akialis K, Tyler J, Vasquez P, Yeh E, Bloom K. Microtubule dynamics drive enhanced chromatin motion and mobilize telomeres in response to DNA damage. Mol Biol Cell. 2017;28:1701–1711. doi: 10.1091/mbc.E16-12-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 134.Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tüysüz B, Nürnberg G, Kiess W, Koegl M, Baessmann I, Buruk K, Toraman B, Kayipmaz S, Kul S, Ikbal M, Turner DJ, Taylor MS, Aerts J, Scott C, Milstein K, Dollfus H, Wieczorek D, Brunner HG, Hurles M, Jackson AP, Rauch A, Nürnberg P, Karagüzel A, Wollnik B. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dinçer T, Yorgancıoğlu-Budak G, Ölmez A, Er İ, Dodurga Y, Özdemir ÖM, Toraman B, Yıldırım A, Sabir N, Akarsu NA, Semerci CN, Kalay E. Analysis of centrosome and DNA damage response in PLK4 associated Seckel syndrome. Eur J Hum Genet EJHG. 2017;25:1118–1125. doi: 10.1038/ejhg.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]