Abstract
Protein homeostasis, or proteostasis, is essential for cell function, development, and organismal viability. The composition of the proteome is adjusted to the specific requirements of a particular cell type and status. Moreover, multiple metabolic and environmental conditions challenge the integrity of the proteome. To maintain the quality of the proteome, the proteostasis network monitors proteins from their synthesis through their degradation. Whereas somatic stem cells lose their ability to maintain proteostasis with age, immortal pluripotent stem cells exhibit a stringent proteostasis network associated with their biological function and intrinsic characteristics. Moreover, growing evidence indicates that enhanced proteostasis mechanisms play a central role in immortality and cell fate decisions of pluripotent stem cells. Here, we will review new insights into the melding fields of proteostasis and pluripotency and their implications for the understanding of organismal development and survival.
Keywords: Autophagy, Chaperones, Differentiation, Pluripotency, Proteostasis, Proteasome, Stress responses
Introduction
Since proteins participate in almost every biological process, protein homeostasis (or proteostasis) is essential for cell function, development, and organismal viability. Typically, the proteome of a mammalian cell contains 10,000 to 20,000 different proteins [1]. Many parameters determine the integrity and quality of the proteome such its composition, the cellular localization of every single protein as well as protein–protein interactions. The proteome is tightly regulated in a dynamic process adjusted to the specific requirements of a particular cell type and status. Moreover, multiple metabolic and environmental conditions challenge the integrity and functionality of the distinct proteins that compose the proteome [2]. A complex network of integrated and competing cellular mechanisms regulates the concentration, folding, trafficking, and interaction of proteins [3, 4]. As such, this proteostasis network monitors proteins from their synthesis through their degradation. The human proteostasis network consists of about ~1400 proteins including regulatory factors and components of macromolecular machineries involved in the synthesis, folding, and degradation of proteins [2, 5].
Under physiological conditions, the proteostasis network is adjusted with high versatility in response to distinct stimuli [3–5]. During development, cell differentiation triggers a myriad of changes in the composition of the proteome. Therefore, proteostasis determines successful organismal development [6]. Furthermore, the proteostasis network is rewired in response to different environmental challenges and pathological conditions such as oxidative or thermal stress and misfolding-prone mutations [5, 7]. Hence, the survival of an organism is determined by its ability to preserve the quality of the proteome [5]. With age, somatic cells lose extensive command of proteostasis: widespread, dysregulation of protein function, aberrant changes in protein synthesis, a generalized down-regulation of chaperones, and impairment of proteolytic machineries often appear in post-mitotic cells as well as adult stem and progenitor cells across time [5–10]. This decline in proteostasis contributes to the characteristic loss of tissue homeostasis of aging organisms [11]. Moreover, proteostasis dysfunction is linked with multiple age-associated disorders including Alzheimer’s, Huntington’s, or Parkinson’s disease and cancer [12]. Conversely, mechanisms that preserve or enhance the proteostasis network until late in life extend healthspan and delay the onset of age-related disorders [5, 7, 13].
While somatic stem cells lose their ability to maintain proteostasis with age [8–10], immortal pluripotent stem cells exhibit a tightly regulated proteostasis network associated with their biological function and intrinsic characteristics [6]. The gold standard of pluripotency is the embryonic stem cell (ESC), which is derived from the inner cell mass of early-stage preimplantation embryos [14]. Alternatively, somatic cells can be reprogrammed by ectopic expression of transcription factors to generate induced pluripotent stem cells (iPSCs) that share similar characteristics with ESCs [14, 15]. Both ESCs and iPSCs replicate indefinitely without undergoing senescence while maintaining their pluripotency [16]. While the transcriptional, epigenetic, and signaling regulators of ESC/iPSC identity have been a primary focus of research efforts, growing evidence indicates that proteostasis regulatory mechanisms also play a central role in ESC/iPSC self-renewal, pluripotency, and cell fate decisions [17–19]. Defects in proteostasis lead to the accumulation of damaged proteins that could result in impairment of ESC pluripotency and immortality. In addition, the passage of these aberrant proteins to progenitor cells during asymmetric divisions may compromise organismal development and aging. As the origin of multicellular organisms, pluripotent stem cells must have stringent mechanisms to protect their proteome from any proteostatic imbalance that would otherwise compromise their function and immortality [6]. Here, we will review new insights into the intrinsic regulation of proteostasis of pluripotent stem cells. Moreover, we will discuss the implications of these findings for the understanding of cell differentiation as well as organismal development and survival.
Protein synthesis
Protein translation is a fundamental step of gene expression by which ribosomes decode mRNAs to synthesize new proteins as linear chains of amino acids [1]. Protein translation consists of three phases: initiation, elongation, and termination. Regulation of protein synthesis can occur at any phase of translation and it may be either specific for distinct transcripts or specific for global. For instance, phosphorylation of the translation initiation factor eIF2Aα impairs the recruitment of the Met-tRNA to the 40S ribosome subunit, resulting in down-regulation of global translational rates [20, 21]. Protein translation is differentially regulated among distinct cell types, a process that contributes to establish and maintain differences in cell identity and function [22].
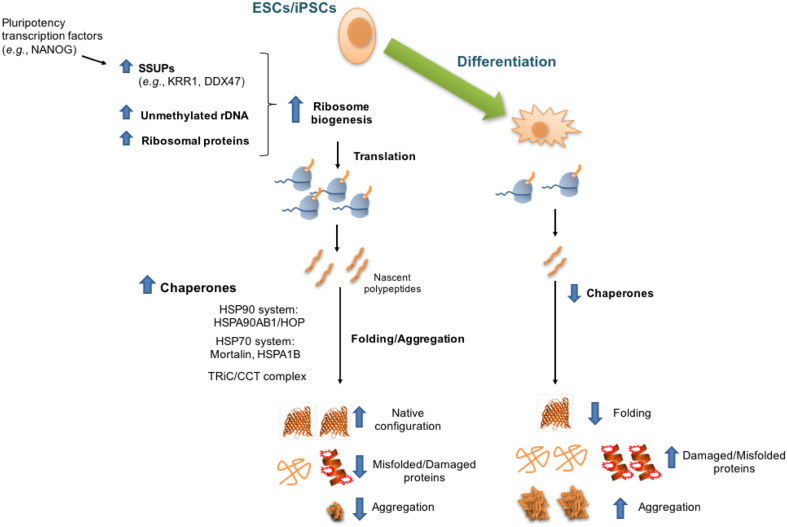
Given that protein synthesis rates impinge upon multiple biological processes that determine cell identity and status, this process is tightly regulated. For instance, the balance between cell proliferation and differentiation crucially depends on activation or repression of protein translation [22]. Several studies in animal models demonstrate that changes in global translational rates are essential to facilitate transition into differentiated states. This is the case of mouse haematopoietic stem cells, which require enhanced protein synthesis to maintain their stemness [23]. In Drosophila melanogaster and Caenorhabditis elegans, precise regulation of translational rates influences the function and proliferation of immortal germline stem cells (GSCs), which are designed to generate gametes for reproduction and maintain an unlimited proliferative capacity [24–27]. Given their biological function and particular characteristics, ESCs must have intrinsic regulatory mechanisms to control protein translation (Fig. 1). Emerging evidence indicates that ESCs exhibit high demand for increased translational rates to sustain their elevated self-renewal activity and pluripotency [19]. In mouse ESCs (mESCs), down-regulation of global protein synthesis rates results in loss of pluripotency as observed with either translational inhibitors or knockdown of genes involved in ribosome biogenesis [19]. Moreover, this study showed that the high global translational rates of mESCs decrease during their differentiation into either embryoid bodies or neural cells [19]. General up-regulation of translation in mESCs is linked with their elevated expression of several subunits (e.g., Ddx47, Krr1) of the small subunit processome (SSUP), a complex that mediates 18S rRNA biogenesis and intensify global protein synthesis [19]. Concomitantly, knockdown of distinct SSUP components impairs 40S ribosome biogenesis in mESCs. Pluripotency factors such as Nanog promote the high expression of SSUP subunits in mESCs, providing a direct link between pluripotency and elevated protein synthesis [19]. In addition, the translation of ribosomal proteins itself undergoes a strong repression when mESCs differentiate into embryoid bodies [28]. While ribosomal protein genes are highly expressed in embryoid bodies, their transcripts are translated less efficiently than the typical mRNAs. In support of a link between enhanced protein synthesis and pluripotency, mESCs produce high levels of pre-rRNA by maintaining the rDNA promoter in an unmethylated state [29]. Whether the aforementioned regulatory mechanisms are evolutionary conserved in human ESCs (hESCs) and iPSCs remained to be elucidated. However, it is important to note that other findings do not support a model of increased global translational rates in ESCs. A previous study indicated that protein translation is parsimonious in mESCs, whereas the global loading of mRNAs in translating polysomes increases during early differentiation into embryoid bodies, resulting in increased protein synthesis and content [30]. This process is modulated by increased phosphorylation of 4EBP1 through activation of the mTOR pathway in embryoid bodies, resulting in the release of active eIF4E complexes that up-regulate global translation [30]. Moreover, mESCs also exhibit an intrinsic translation of upstream open reading frames (uORFs) by which these cells precisely regulate protein synthesis to impact gene expression genome-wide. uORFs are central regulators of gene expression located in the 5′UTR of the mRNA and, therefore, precede the initiation codon of the main codon region [31]. When translation starts from the uORF, protein synthesis of the actual ORF is impaired [31]. Interestingly, mESCs display increased uORF translation compared with their differentiated counterparts [28]. Given that approximately, half of the human transcripts possess uORFs [31], this process could also play an important role in the regulation of global translational rates of hESCs.
Fig. 1.
Intrinsic regulation of proteostasis-related anabolic processes of pluripotent stem cells. Scheme showing specific components of protein translation and folding nodes, which are differentially expressed in pluripotent stem cells compared with their differentiated counterparts. Pluripotent stem cells exhibit up-regulated global levels of protein synthesis rates, a process modulated by enhanced expression of several subunits of the small subunit processome (SSUP), unmethylated state of rDNA promoter, and translation of ribosome subunits. To assist the folding of high amounts of newly synthesized proteins, pluripotent stem cell function requires increased expression of chaperone core machinery genes. In addition, the intrinsic chaperome network of pluripotent stem cells maintains the proper folded state of proteins from their synthesis through their degradation, resulting in decreased accumulation of damaged/misfolded proteins and aggregates. Although differentiated cells exhibit decreased translational rates, a concomitant down-regulation in the chaperone/folding system diminishes their ability to refold damaged/misfolded proteins and restrain protein aggregation
Remarkably, post-transcriptional modifications of RNA modulate ESC identity and cell fate decisions. N6-methyladenosine (m6A), the most abundant post-transcriptional modification in RNA [32], is involved in all aspects of RNA metabolism, including mRNA turnover [33, 34] and translation efficiency [35]. Whereas several studies propose that m6A modifications are required to maintain ESC identity [17, 34], other findings suggest that m6A determines ESC commitment to differentiated states [35, 36]. Notably, the chromatin-associated zinc finger protein 217 (ZFP217) modulates the global levels of m6A in ESCs. ZFP217 interacts with epigenetic modulators to activate the transcription of multiple core pluripotency factors while reduces m6A deposition on these pro-pluripotency transcripts to promote their translation [17]. Likewise, growing evidence indicates that distinct RNA-binding proteins (RBPs) also play central roles in hESC identity and cell fate decisions. RBPs participate in practically any step of gene expression involving RNA such as turnover and translation [18]. One of the most studied RBPs in the context of pluripotency is LIN28, which is highly expressed in ESCs to regulate the stability and translation of hundreds of mRNAs that define ESC identity [37].
Taken together, global protein synthesis rates as well as post-transcriptional co-regulation of functionally related genes determine ESC identity. However, there are still many intriguing aspects to be examined in the field. Moreover, most of the studies used mESCs as a primary model, whereas translational regulatory mechanisms remain largely unknown in human pluripotent stem cells. Thus, it will be particularly fascinating to define how human pluripotent stem cells regulate global translational rates and their changes during cell differentiation/reprogramming.
Protein folding and aggregation
Once proteins are synthesized in the ribosome, they must fold in a proper three-dimensional structure to perform their specific biological function [2]. Protein folding is facilitated by molecular chaperones [38]. Besides altered protein activity, dysfunction of nascent protein folding can also result in aberrant aggregation [1]. Since chaperones are critical for the folding of numerous nascent proteins, imbalances in the chaperone network can accelerate aggregate formation. Chaperones interfere at different steps of the aggregation process, including primary nucleation, elongation, and fragmentation of fibril and secondary nucleation [1]. In addition, chaperones participate in refolding or scavenging activities when the structure of proteins is challenged by distinct conditions, including high temperatures or oxidative stress as well as the aging process [2, 5, 38]. Thus, molecular chaperones are central components of the proteostasis network that assure the proper folding and localization of proteins through their life cycle [5]. To sustain the integrity of the proteome under both physiological and stress conditions, several chaperone families and co-chaperones cooperate preserving the balance between protein synthesis, folding, and degradation [1]. The human chaperome is formed by 332 chaperones and co-chaperones with specific functions and intracellular localization [39]. These chaperones are grouped in different families such as the 90 kilodalton heat-shock proteins (HSP90s), HSP70s, HSP60s (also known as chaperonins), HSP40s, and small HSPs [1, 5]. Each of these major chaperone families are essential for cell viability, indicating that they are enrolled in non-overlapping functions [40]. Whereas distinct chaperones are involved in protein biogenesis, other set evolved to essentially protect the cell from proteotoxic stress [41].
Growing evidence indicates that specific chaperones have a role in pluripotency and differentiation (Fig. 1). In ESCs, the intense quality control of the proteome by the chaperome network is adjusted to their high proliferative and translational rates as well as the need to maintain a pristine proteome that can be transferred to their differentiated counterparts [6]. Indeed, ESCs have an intrinsic chaperome network with high expression of multiple chaperones, whereas down-regulation of these chaperones leads to ESC differentiation [42, 43]. Moreover, this intrinsic chaperome network could contribute to explain why ESCs do not age [6]. In the following sections, we will discuss recent insights into the link between chaperones, proteotoxic stress resistance, and pluripotency.
HSP90s
HSP90s are involved in many nodes of proteostasis such as proper protein folding, stabilization of proteins against heat stress, and degradation of misfolded proteins. HSP90s form homodimers that cooperate with over 20 co-chaperones and regulators [e.g., Cdc37, AhaI, p23/SbaI, or the HSP organizing protein (HOP)] that modulate their ATPase activity, recruit clients, and facilitate client maturation [1, 5, 44]. In humans, there are five functional HSP90 isoforms. HSP90B1 (also known as GRP94) and TRAP1 are essentially located in the ER and mitochondria, respectively [45, 46]. In the cytosol, there are two types of HSP90s: the constitutively expressed HSP90AB1 and the stress-induced HSP90AA1 and HSP90AA2 [47].
Highly proliferative cells such as cancer cells exhibit increased levels of cytosolic HSP90 [48]. Likewise, mESCs and hESCs express high levels of cytosolic HSP90 as well as its co-chaperone HOP [42, 43]. Down-regulation of HSP90 activity by either specific inhibitors or knockdown leads to ESC differentiation [49]. The increased levels of cytosolic HSP90 and co-chaperones not only confer enhanced stress tolerance to ESCs but also regulate key pluripotency components [43]. For instance, cytosolic Hsp90 binds Oct4 and Nanog in mESCs, protecting these pluripotency factors from degradation by the proteasome [49]. In addition, Hsp90 and Hop play a role in the regulation of the JAK/STAT3 signaling pathway [50, 51], which is essential for maintenance of mESC cultures in the absence of feeder cells [14]. Both cytosolic Hsp90 and Hop are required for the activation of Stat3 phosphorylation and its nuclear translocation [50, 51]. Notably, the leukaemia inhibitory factor (LIF), a key ingredient for culturing mESCs that activates the JAK–STAT3 pathway [14], promotes the interaction of Hsp90 with Stat3 during self-renewal [51].
The role of GRP94, the primary HSP90 for the folding of secretory and membrane proteins in the ER, has also been examined in the context of ESC pluripotency. Grp94 −/− mouse embryos die on day 7 of gestation and fail to develop mesoderm and primitive streak [52]. Although Grp94 −/− mESCs can proliferate in culture and differentiate into all the three germ layers, they cannot further differentiate into cardiac, smooth, or skeletal muscle [52]. Moreover, differentiated cells from Grp94 −/− mESCs are hypersensitive to serum deprivation and show impaired secretion of growth factors such as the insulin-like growth factor II [52].
HSP70s
HSP70 family of chaperones is involved in many aspects of cellular proteostasis, including protein synthesis, translocation, aggregation, and degradation [1, 53, 54]. Multiple co-chaperones interact with HSP70s to assist them in their vast range of proteostatic tasks. The most important co-chaperones are DNAJ-domain-containing HSP40s [39, 55] and nucleotide exchange factors (NEFs) such as BAG1 [54]. De novo folding is modulated by the combined action of HSP70s with different co-chaperones and other molecular chaperones including HSP90s and chaperonins. Once HSP70 is loaded with a client, it couples with HSP90 via HOP or interacts directly with chaperonins to deliver the client for further folding [1]. In stressed cells, the presence of DNAJ-domain-containing HSP40s co-chaperones and ATP potentiates HSP70 chaperone efficiency [56]. Cytosolic HSP70 is not only required for proper protein folding, but also for 26S proteasome-dependent degradation of misfolded proteins distributed in the cytosol itself or extracted from the endoplasmic reticulum (ER) and mitochondria. Moreover, HSP70 can also target misfolded proteins to the lysosome in a process known as chaperone-mediated autophagy (CMA) [53, 57]. In addition, HSP70s also contribute to the clearance of protein aggregates and their degradation to peptides [53]. Accordingly, overexpression of HSP70s can protect from age-and disease-associated tissue degeneration in brain, heart, and skeletal muscle [58]. Conversely, dysfunction of HSP70 activities can lead to severe human degenerative diseases such as Alzheimer’s or Parkinson’s and type 2 diabetes.
The distinct members of the HSP70 family show differential intracellular location. HSPA8 is the major HSP70 expressed in the cytosol, whereas HSPA5 (BIP) and HSPA9 (mortalin) are the most abundant HSP70 species in ER and mitochondria, respectively [53]. Interestingly, immunocytochemical analyses showed that HSPA8 can be detected on the surface of hESCs, whereas this surface expression is markedly decreased during differentiation, suggesting that HSPA8 is a putative cell-surface marker for undifferentiated hESCs [59]. However, it is unknown whether surface expression of HSPA8 impinges upon ESC function and cell reprogramming. Although the surface expression of HSPA8 is down-regulated during neural differentiation [59], it is important to note that the total levels of HSPA8 do not change upon neuroectodermal differentiation [42]. In mESCs, the levels of other cytosolic HSP70s (i.e., Hspa1a and Hspa1b) are increased when compared with murine differentiated cells [60]. Likewise, hESCs also exhibit increased expression of HSPA1B [61]. Although the impact of these increased levels of specific cytosolic HSP70s has not been examined, their cellular functions suggest a role in the regulation of intrinsic characteristics of pluripotent stem cells. For instance, increased HSP70s may be necessary to cope with the high protein-folding requirements originated from enhanced protein synthesis and proliferation rates. Moreover, HSPA1B confers resistance to apoptosis [62], and therefore, this chaperone could contribute to the increased resistance of ESCs compared with differentiated cells [60]. Given that a small number of pluripotent stem cells give rise to all the organismal tissues, another possibility is that increased HSP70 levels confer resistance to various stress (including proteotoxic and oxidative stress) assuring the generation of healthy differentiated cells. In these lines, the levels of mortalin, the mitochondrial HSP70, are also increased in ESCs [60, 63]. Mortalin is involved in multiple biological processes such as control of cell proliferation [64] or stress response [65]. Overexpression of mortalin in somatic cells confers survival advantage to these cells and assists telomerase in cell immortalization [66], factors that have a key role in maintenance of ESC identity.
HSP60s (chaperonins)
Chaperonins exhibit ATPase activity to bind and facilitate the folding of nascent or misfolded proteins. The 60 kDa chaperonin family of proteins consists of both constitutively expressed and stress inducible members [39]. These chaperonins can assemble into multi-subunit protein-folding complexes that are divided into two major groups: (1) Group I chaperonins, which are present in bacteria and organelles of endosymbiotic origin (i.e., chloroplasts and mitochondria) and (2) Group II chaperonins (TRiC/CCT), which are found in the cytosol of eukaryotes and in Archea [67]. Notably, pluripotent stem cells exhibit high levels of HSPD1 [42, 68], a mitochondrial chaperonin that not only participates in the transport and folding of imported proteins but also promotes refolding of misfolded proteins ensued from stress conditions in the mitochondrial matrix [69]. Moreover, hESCs also exhibit increased levels of HSPE1 [42], a co-chaperone of HSPD1 [70]. These up-regulated levels of both HSPD1 and HSPE1 decrease upon differentiation [42].
In addition, hESCs and iPSC exhibit intrinsic up-regulated levels of the eukaryotic cytosolic TRiC/CCT chaperonin complex [42]. TRiC/CCT consists of two-stacked rings that form a cylindrical structure with a central cavity to assist the folding of non-native proteins [71]. Each ring of the TRiC/CCT complex is formed by eight paralogous subunits that have different client recognition specificities [72]. TRiC/CCT is essential for cell viability and facilitates the proper folding of approximately 10% of the nascent proteome [67, 73]. For instance, TRiC/CCT complex assists the folding of key cytoskeletal proteins (e.g., actin and tubulin) maintaining cell morphology and allowing for cell division [5]. Human pluripotent stem cells exhibit an increased expression of several CCT subunits (e.g., CCT3, CCT4, and CCT8) compared with their differentiated counterparts, resulting in enhanced levels of the TRiC/CCT complex [42]. Conversely, TRiC/CCT assembly decreases during differentiation. Notably, this decline is already significant in multipotent cells such as neural progenitor cells before terminal differentiation [42]. On the contrary, pluripotent stem cells maintain high levels of CCT subunits after numerous passages, further supporting a link between pluripotency status and enhanced expression of CCTs [42]. Indeed, a mild knockdown of different CCT subunits decreases the levels of pluripotency markers in hESCs/iPSCs, whereas strong CCT down-regulation induces cell death and detachment of hESC/iPSC colonies [42]. The mechanisms by which increased assembly of the TRiC/CCT complex maintains pluripotency are unknown. Enhanced TRiC/CCT assembly may be required for the folding of central regulatory or structural proteins involved either in maintenance of pluripotency or generation of healthy differentiated cells. Interestingly, a study performed in calreticulin−/− mESCs showed down-regulation of Cct2, Cct3, and Cct7 subunits in these cells [74]. Calreticulin, a chaperone that binds to misfolded/damaged proteins preventing their export from the ER, is essential for cardiac development in mice [75] and proper myofibril formation during cardiomyocyte differentiation of mESCs [76]. Given that TRiC/CCT modulates folding and actin dynamics, the decrease of Cct subunits in calreticulin−/− mESCs may forecast the myofibrillar disarray reported in their cardiomyocytes counterparts [74].
Besides assisting the folding of newly synthesized proteins [77], TRiC/CCT complex also regulates the aggregation and toxicity of misfolded proteins [78]. For instance, TRiC/CCT modulates the aggregation of mutant huntingtin (HTT) [79–81], the protein underlying Huntington’s disease [82]. Accordingly, dysfunction of the TRiC/CCT complex induces aggregation of mutant HTT and worsens Huntington’s disease-related changes in yeast, C. elegans, and mammalian models [79–81]. In addition, TRiC/CCT also hampers amyloid fibril formation of α-synuclein A53T (one of the most studied mutations linked with Parkinson’s disease) suggesting a general function of TRiC/CCT in controlling amyloidopathies [83]. Thus, increased TRiC/CCT levels could function in pluripotent stem cells to maintain an intact proteome for either self-renewal or the generation of progenitor cells. Indeed, enhanced TRiC/CCT assembly is required for the striking ability of pluripotent stem cells derived from Huntington’s disease patients to maintain proteostasis of aggregation-prone mutant HTT [42]. Since the levels of CCT subunits further decrease in somatic tissues with age [39], a detailed study of how pluripotent stem cells sustain increased TRiC/CCT assembly could have important implications for correcting proteostasis defects in age-related diseases. In particular, CCT8 subunit has been established as a key promoter of TRiC/CCT assembly in pluripotent stem cells [42]. Remarkably, mimicking proteostasis of iPSCs in post-mitotic somatic tissues by ectopic expression of CCT8 is sufficient to increase TRiC/CCT assembly. Moreover, somatic overexpression of CCT8 delays the age-associated proteostasis demise and extends longevity in C. elegans. Notably, CCT8 also reduces toxic protein aggregation and ameliorates Huntington’s disease-related changes in C. elegans models [42]. One step further will be to define how the levels of CCT8 and other CCT subunits are modulated in pluripotent stem cells with the aim to mimic this regulation in somatic tissues of disease models.
HSP40s
HSP40s play a role as holding chaperones and HSP70 co-chaperones, allowing for substrate targeting of HSP70 family members [39]. Although HSP40s lack ATPase activity, they can stimulate ATP hydrolysis exerted by HSP70s during protein folding and disaggregation activities [84]. The levels of several DNAJ-domain-containing HSP40s such as DNAJA1, DNAJA2, DNAJB1, DNAJC7, DNAJC8, and DNAJC9 decrease during neuronal differentiation of hESCs [42]. However, whether increased levels of these HSP40s are required for hESC self-renewal, pluripotency and/or differentiation remain to be elucidated.
Small HSPs
One of the main functions of small HSPs is to prevent the accumulation and aggregation of misfolded proteins by facilitating their refolding or degradation [85]. Although the impact of small HSPs has not been studied in a comprehensive manner in the context of pluripotency, cumulative evidence indicates that these chaperones could play important roles in pluripotent stem cells. Several studies have shown changes in the levels of specific small HSPs (e.g., HSP27) during differentiation [42, 60, 86]. In particular, Hsp27 levels increase during the early stages of differentiation [86]. Notably, this up-regulation of Hsp27 prevents differentiating mESCs from undergoing apoptosis and, therefore, allows for their differentiation into embryoid bodies [86].
Protein clearance systems
Protein degradation systems maintain the proper concentration of many regulatory factors and adjust their levels in response to intracellular signaling pathways and external stimuli [7]. Moreover, proteolytic systems terminate damaged and misfolded proteins when refolding mechanisms are not sufficient to assure proper protein function, reducing proteotoxicity stress and protein aggregation [7]. Different proteolytic systems such as the ubiquitin proteasome system (UPS), autophagy, calpains, and caspases contribute to clearance of proteins [7]. Among them, intrinsic UPS and autophagy mechanisms have been linked with pluripotency, differentiation, and cell reprogramming [87–89].
The ubiquitin proteasome system (UPS)
The UPS is the major selective proteolytic system in eukaryotic cells [90]. As such, the UPS determines half-life of numerous proteins and prevents accumulation of damaged proteins and aggregates, regulating multiple biological processes [4]. The first step of the UPS-mediated degradation is the sequential attachment of ubiquitin molecules to the target protein (Fig. 2). This process is accomplished through a three-step cascade mechanism [91]. First, ubiquitin is activated in an ATP-dependent manner by the ubiquitin-activating enzyme (E1) [92]. The activated ubiquitin is transferred to ubiquitin-conjugating enzymes (E2s) via formation of E2-ubiquitin thioester structure [93]. Then, E3 ligases catalyze the attachment of ubiquitin to their specific target proteins by binding both the E2-ubiquitin thioester structure and the target protein [92]. The same sequential mechanism links additional molecules to the primary ubiquitin via internal ubiquitin lysines, forming a polyubiquitin chain. Given that the proteasome regulates the abundance of pluripotency factors such as OCT4 or NANOG, the UPS is involved in maintenance of hESC identity and differentiation [94]. Moreover, the UPS also have a critical role in cell reprogramming [87]. ESCs exhibit and intrinsic UPS network characterized by differential expression of proteasome regulators (Fig. 2) and E3 ligases compared with differentiated cells [87, 89]. E3 ligases are responsible for substrate selection providing specificity to the proteolytic process. While there are only 2 E1 and 35 E2s, over 600 E3 ligases have been identified in humans so far. Among them, specific E3s regulate pluripotency and differentiation (recently reviewed in [95]). For instance, the E3 ligase Huwe1 polyubiquitinates N-myc (an essential transcription factor for ESC identity) and promotes its degradation by the proteasome, allowing for neural differentiation of mESCs [96].
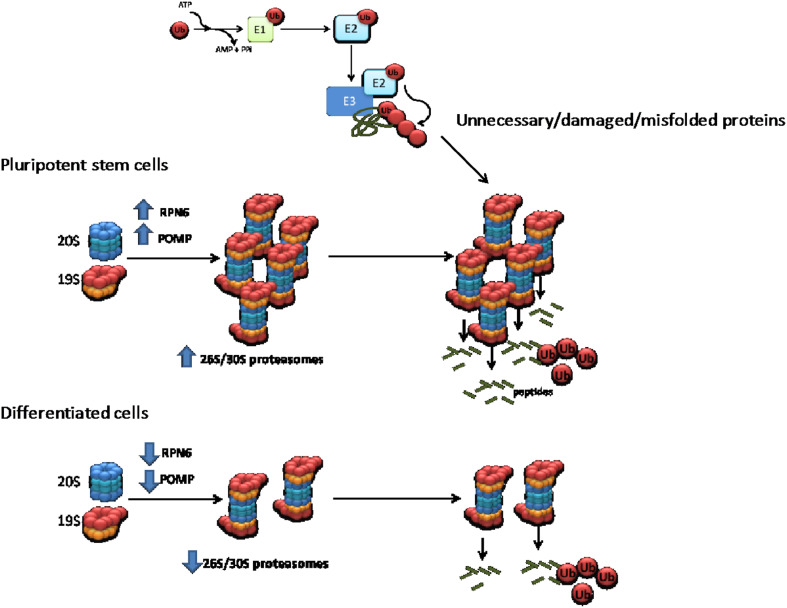
Fig. 2.
Intrinsic regulatory mechanisms of 26S/30S proteasome activity in pluripotent stem cells compared with their differentiated counterparts. Proteasome-mediated degradation starts with the sequential attachment of ubiquitin molecules to the target substrate (e.g., regulatory, unnecessary, damaged, and misfolded proteins), a process mediated by a three-step cascade mechanism. First, ubiquitin is activated in an ATP-dependent manner by the ubiquitin-activating enzyme (E1). The activated ubiquitin is transferred to ubiquitin-conjugating enzymes (E2s) via formation of E2-ubiquitin thioester structure. Then, E3 ligases catalyze the attachment of ubiquitin to their specific target by binding both the E2-ubiquitin thioester structure and the substrate protein. The same sequential mechanism links additional molecules to the primary ubiquitin through internal ubiquitin lysines, forming a polyubiquitin chain. E3 ligases provide specificity to the proteasomal-degradation process and over 600 E3 ligases have been identified in humans so far. After the polyubiquitination cascade process, target proteins are recognized and degraded by the 26S/30S proteasome. Active 26S/30S proteasomes are formed by the interaction between the 20S catalytic core and the 19S regulatory particle. Pluripotent stem cells exhibit increased levels of RPN6 and POMP resulting in increased assembly and activity of the 26S/30S proteasome. On the contrary, the levels of RPN6 and POMP are down-regulated in differentiated cells resulting in decreased assembly of 26S/30S proteasomes
After the polyubiquitination cascade process, target proteins can be recognized and degraded by the 26S/30S proteasome [97, 98]. The 26S/30S proteasome is formed by the interaction of the 20S catalytic core with the 19S regulatory complex [98]. Notably, both hESCs and mESCs exhibit increased 26S/30S proteasome assembly and activity (Fig. 2) [87, 89]. Although ESCs have enhanced proteasome activity, these cells are extremely more sensitive to proteasome inhibition compared with differentiated cells [89, 99]. Moreover, elevated proteasome activity is required for ESC function as a mild down-regulation of the proteasome results in decreased levels of pluripotency factors (e.g., OCT4, NANOG) and concomitant differentiation [89, 99]. In hESCs and iPSCs, increased proteasome activity is induced by high expression of PSMD11/RPN6 [89], a 19S scaffolding subunit that also promotes the interaction between the 19S regulatory complex and the 20S catalytic core [100]. Interestingly, increased levels of PSMD11/RPN6 in immortal hESCs/iPSCs are regulated by FOXO4, a forkhead transcription factor associated with organismal longevity. This modulation of proteasome activity by FOXO4 is necessary for ESC differentiation into neural cells [101]. Besides PSMD11/RPN6, other proteasome components and regulators are also increased in ESCs. For instance, ESCs exhibit up-regulation of the deubiquitinating enzyme Psmd14 [87] and the proteasome maturation protein (POMP) [102]. POMP is a proteasome chaperone that not only maintains proliferation of hESCs but also regulates cell reprogramming [102]. Remarkably, POMP levels in ESCs are regulated by NRF2, a transcription factors that confers stress tolerance [102].
Besides its role in regulating the concentration of pluripotency/differentiation factors, increased proteasome activity could also terminate damaged proteins and maintain an intact proteome in ESCs. In support of this hypothesis, overexpression of PSMD11/RPN6 in somatic tissues increases stress tolerance and ameliorates the accumulation of protein aggregates in Huntington’s disease models [103]. Moreover, a role of enhanced proteasome activity in degradation of damaged proteins has been established in differentiating mESCs [104, 105]. At this stage, a variant of the proteasome known as immunoproteasome acquires a key relevance. The immunoproteasome, normally associated with antigen-presenting cells, is generated by replacement of the 20S catalytic subunits by β1i, β2i, and β5i and the 19S regulatory particle by the PA28αβ complex (also known as 11S complex). During the early steps of differentiation, ESCs trigger the termination of damaged proteins by induction of the catalytic subunit β5i (PSMB8) and the immunoproteasome regulatory activator PA28αβ [104, 105].
Autophagy: the bulk degradation pathway
Autophagy is a self-catabolic process that degrades cytosolic fractions as well as damaged or outlived organelles and macromolecules. Autophagy is essential in biological processes that require extensive cellular restructuration, such as embryogenesis, cellular differentiation, or reprogramming [9, 106–109]. Lysosome, the catalytic component of the autophagy process, contains a variety of cellular hydrolases, lipases, nucleotidases, glycosidases, as well as proteases with the highest activity at acidic pH. Autophagy is emerging as a selective mechanism of protein degradation [110]. Specific adaptors such as p62 (SQSTM1) recognize ubiquitinated proteins and target these substrates to the autophagy machinery [110]. Lysosomal proteolysis results in free amino acids, small di- and tri-peptides as well as larger peptides that are released into the cytosol to be either further metabolized to obtain energy or recycled to synthesize de novo molecules [107, 111]. The autophagy process does not only function as a cellular recycling pathway but also preserves cellular homeostasis acting as a quality control mechanism of proteins and organelles. As such, autophagy also contributes to scavenge misfolded and aggregated proteins [5, 7]. Whereas protein aggregates can block the proteasome machinery [112], autophagy is able to degrade large protein complexes or aggregates [5, 7]. This function includes degradation of aberrant aggregates formed by neurodegenerative-associated proteins such α-synuclein, tau and polyglutamine-expanded proteins (e.g., mutant HTT) [113]. Accordingly, dysregulation of this process is associated with age-related disorders such as Parkinson’s, Alzheimer’s and Huntington’s disease [7, 11].
Depending on the cargo recognition system and the mechanism of delivery to lysosomes, autophagy is classified into different modalities [114]. The best characterized types of autophagy are macroautophagy, microautophagy, and CMA. In microautophagy, parts of the cytoplasm are directly engulfed by the lysosome membrane [115]. In CMA, cytoplasmic proteins are recognized by chaperones such as HSP70 through a consensus sequence and transferred to the lysosome for their proteolysis [116]. Macroautophagy (hereafter referred as autophagy) is an evolutionary conserved process characterized by the engulfment of organelles or cytosolic regions by a double membrane vesicle known as autophagosome, which then fuses with the lysosome leading to degradation of its contents [115]. Autophagy and microautophagy can degrade both organelles and proteins, whereas CMA exclusively participates in protein termination [116].
Autophagy is based on the orchestrated action of many proteins and protein complexes. The integral part of the autophagy process is regulated by the autophagy-related (ATG) genes, which were first characterized in yeast as essential for nutrient stress and starvation, but have homologues in almost all other organisms, including mammals [109, 117]. Autophagy starts with the formation of a double membrane structure known as the phagophore, a process regulated by the ULK complex (consisting of ULK1, ATG13, FIP200, and ATG101) [7] (Fig. 3). Then, the VPS34–BECN1 complex (composed of VPS34, BECN1, AMBRA1, and ATG14L) promotes the expansion of the phagophore. Once the cytoplasmic fraction is engulfed into the phagophore, the membrane elongates until it closes forming the autophagosome. Both elongation and closure of the autophagosome membrane are regulated by the ATG12–ATG5–ATG16L1 complex, which enhances the conjugation of cytosolic LC3 (LC3I) to phosphatidylethanolamine. LC3–phosphatidylethanolamine conjugate (LC3II) is recruited to the membrane, and then, LC3II–positive autophagosomes are trafficked to the lysosome for degradation. Autophagy is modulated by many signaling pathways such as Sirtuin 1 (SIRT1), adenosine monophosphate-activated protein kinase (AMPK), and mTOR, a serine/threonine kinase which exists in two complexes named mTORC1 and mTORC2 [118–120]. mTORC1, the mammalian target of rapamycin, modulates the assembly of the ATG complex required for the formation of the autophagosome [121].
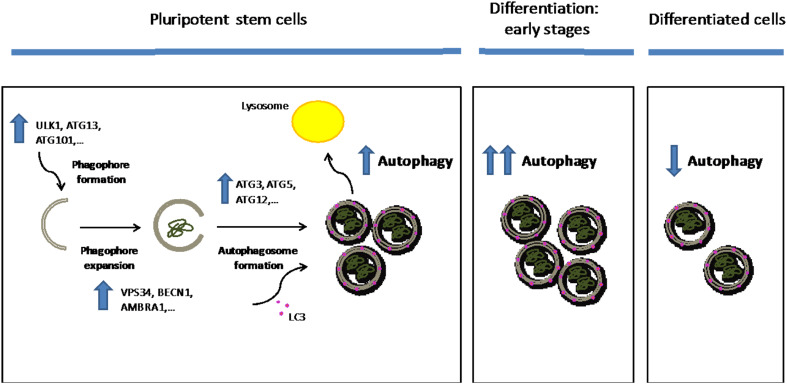
Fig. 3.
Autophagic flux in pluripotency and differentiation. The ULK complex (ULK1, ATG13, FIP200, and ATG101) regulates the formation of the double membrane structure known as the phagophore. Then, the VPS34–BECN1 complex (formed by VPS34, BECN1, AMBRA1, and ATG14L) mediates the expansion of the phagophore. Once the cytoplasmic fraction is engulfed into the phagophore, the membrane elongates until it closes forming the autophagosome, a process regulated by the ATG12–ATG5–ATG16L1 complex, which enhances the conjugation of cytosolic LC3 (LC3I) to phosphatidylethanolamine. LC3–phosphatidylethanolamine conjugate (LC3II) is recruited to the membrane and then LC3II-containing autophagosomes are trafficked to the lysosome for their degradation. Pluripotent stem cells exhibit a higher basal autophagic flux compared to terminally differentiated cells such as fibroblast and neurons. These up-regulated levels of autophagic flux are sustained by increased expression of core genes involved in different steps of autophagy. Moreover, autophagy–lysosome activity is further up-regulated during early differentiation steps. Once the cells are differentiated, they exhibit decreased autophagic flux compared with undifferentiated pluripotent stem cells
Autophagy is highly up-regulated in oocytes immediately after fertilization [122] and a particular form of autophagy, mitophagy, removes paternal mitochondria from fertilized oocytes [123]. Autophagy is essential for the first stages of embryogenesis (up to the eight-cell stage) for removing old cellular structures and recycling cellular components [109]. Mice carrying an oocyte specific conditional knockout of Atg5 do not proceed the four-to-eight-cell stage if fertilized with Atg5-null sperm. Becn1 −/− and Ambra1 −/− mice also fail to develop [109]. The impact of autophagy in later stages of embryogenesis remains unclear. Knockout mouse models for different Atg genes (such as Atg7 −/−, Atg9 −/−, Atg3 −/−, Atg16L1 −/−, Atg5 −/−) give birth to litters at mendelian frequencies, although they exhibit reduced weight at birth and die within 1–2 days postnatally, presumably due to suckling defects associated with neurological abnormalities [109, 124]. It remains to be elucidated whether other mechanisms such as the UPS can compensate for autophagy dysfunction in the later developmental stages [115].
In vitro, autophagy is also required for ESC differentiation as loss of ATG5 and BECN1 leads to formation of defective embryonic bodies [125]. Moreover, autophagy is essential for mitochondrial homeostasis during pluripotency acquirement and maintenance [126]. A recent study has shown that mESCs exhibit a higher basal autophagic flux when compared with terminally differentiated cells such as neurons or fibroblasts [88]. This high autophagic flux results in increased degradation of long-lived proteins [88]. The powerful autophagic infrastructure of ESCs is maintained by enhanced expression of core genes involved in autophagy initiation (e.g., Ulk1, Atg13, and Atg101), autophagosome formation (e.g., Vps34, Becn1, Ambra1, and Atg14), or ATG12/LC3 conjugation systems (e.g., Atg12, Atg5, Atg3, and Lc3b) [88] (Fig. 3). Likewise, mouse and human iPSCs also exhibit increased levels of basal autophagy and higher expression of key autophagy components such as ULK1/2, BECN1, ATG13, ATG101, ATG12, ATG3, and ATG5 [88, 127]. Interestingly, the transcription factor Foxo1 mediates increased expression of autophagy core machinery genes in ESCs [88]. Conversely, loss of Foxo1 reduces the autophagic flux of ESCs and compromises their self-renewal, pluripotency and differentiation [88]. However, it is important to note that other studies found a further up-regulation of autophagy–lysosome activity in mESCs and hESCs during early differentiation steps [128]. In these lines, both the number of autophagosomes and degradation of p62 markedly increase during neural differentiation. However, these parameters remain identical during the early steps of mesodermal differentiation, indicating differences in autophagy regulation depending on the cell commitment fate [129]. Interestingly, autophagy up-regulation during neural induction inactivates the stress-tolerance transcription factor NRF2 [129], a suppressor of neurogenesis.
Growing evidence indicates that autophagy is essential for cell reprogramming [130–132]. In addition, induction of autophagy enhances reprogramming efficiency [125]. For instance, modulation of mTOR pathway via administration of rapamycin increases reprogramming efficiency by two- to threefold [127]. Interestingly, mTOR is regulated by SOX2, one of the main reprogramming factors. SOX2 can bind directly to the mTOR promoter, leading to derepression of mTOR expression and activation of autophagy [131].
Asymmetric segregation of misfolded proteins
The high proliferation rates invoked by pluripotent stem cells could contribute to their remarkable ability to maintain proteostasis. Asymmetric segregation of damaged proteins was first characterized in the budding yeast and later found evolutionary conserved in mammalian cells. In this process, the mother cell retains a pristine proteome, whereas the daughter cell inherits unnecessary, damaged and misfolded proteins [133–135]. The intermediate filament vimentin provides the physical scaffold for asymmetric inheritance of damaged proteins [133]. An intriguing possibility is that mammalian ESCs promote asymmetric division of damaged proteins to maintain their immortality. In this model, the daughter-differentiating cell would inherit damaged proteins, whereas the mother ESC maintains a pristine proteome. Interestingly, experiments in somatic stem cells support this hypothesis. For instance, embryonic and young mammalian neural stem cells (NSCs) distribute asymmetrically their damaged proteins during cell division [136]. However, this ability decreases with age leading to a more symmetric distribution of damaged proteins between progeny and, therefore, contributing to aging of NSCs pools through organismal lifespan [136]. Whether ESCs induce asymmetric segregation of damaged proteins to maintain their self-renewal and immortality remains to be elucidated.
Proteotoxic stress responses
Under proteotoxic conditions, organisms activate a series of cellular stress responses to correct alterations in proteostasis. Among these mechanisms, the heat-shock response (HSR) can rapidly induce the expression of specific cytosolic chaperones and other proteostasis nodes to overcome drastic temperature changes that challenge the structure of proteins [5, 137]. The HSR protects cells from protein misfolding and ameliorates chronic and acute proteotoxic stress [137]. Interestingly, diminished HSR is associated with neuronal differentiation, resulting in increased vulnerability of neurons to stress-induced pathologies [138]. The transcription factor HSF1 is the master regulator of the HSR. Under proteotoxic conditions such as high temperatures, HSF1 translocates to the nucleus and activates the expression of chaperones such as HSP90s and small HSPs [137]. In normal conditions, loss of HSF1 does not affect hESC function and their differentiation into neural cells [101]. However, it is important to note that human cells express other HSFs (e.g., HSF2) that could compensate the down-regulation of HSF1 levels. Moreover, it will be fascinating to examine whether HSF1 function becomes more relevant in the context of pluripotency under proteotoxic stress conditions.
Besides HSR, other protective mechanisms are activated in response to proteotoxic challenges in specific organelles such as the ER or mitochondria. In the ER, nascent secretory and membrane proteins are folded into their mature structures and enter the cellular vacuolar system. When this process is challenged, the ER unfolded protein response (UPRER) prevents the accumulation of misfolded proteins by three different mechanisms: inhibition of global protein synthesis, increased expression of chaperones to refold misfolded proteins, and activation of degradation of these damaged proteins [139–141]. Cumulative evidence indicates that the UPRER is required for ESC function and differentiation. Induction of the UPRER is necessary for neuronal induction [142], whereas UPRER inhibition halts the differentiation of mESCs by impairing the Smad2 and β-catenin signaling pathways [143]. The UPRER consists of three branches initiated by different misfolding sensors: the activating transcription factor 6 (ATF6), the stress sensors protein kinase RNA-like ER kinase (PERK), and the inositol-requiring protein 1α (IRE1α) [140]. Notably, homozygous deletion of these major UPRER regulators and their downstream effectors (e.g., XBP1) results in organogenesis dysfunction and embryonic lethality [144–147]. The molecular chaperone GRP78/BiP plays a critical role in the activation of the UPRER [140]. In normal conditions, GRP78/BiP binds to the core activators (i.e., IRE1α, PERK, and ATF6) of the UPRER and silence their function. When misfolded proteins accumulate in the ER, they bind to GRP78/BiP resulting in its dissociation from UPRER regulators allowing for their activation [148, 149]. Grp78 −/− mouse embryos do not hatch, fail to grow in culture and exhibit diminished proliferation and increased apoptosis in the inner cell mass, which contain the precursors of ESCs [150]. Thus, these findings indicate that GRP78 is essential for viability of pluripotent cells.
As the ER compartment, a vigilant mechanism formed by chaperones and proteases, known as the mitochondrial UPR (UPRmt), is activated in the mitochondria when the accumulation of unfolded proteins exceeds the folding capacity of the mitochondrial matrix [151]. The impact of the UPRmt on pluripotency and cell fate decisions remains to be examined in detail.
Concluding remarks
A comprehensive understanding of pluripotency, differentiation, and somatic cell reprogramming is necessary for pluripotent stem cells to hold great promise for regenerative medicine. Maintenance of hESC function and cell reprogramming requires the precise coordination of transcriptional factors, epigenetic modifiers, and post-transcriptional regulatory mechanisms. Hence, considerable research efforts have been committed to defining the transcriptional network, chromatin regulators and RNA modifiers of pluripotency. As reviewed here, cumulative evidence indicates that proteostasis is also a core component of the pluripotency regulatory network. ESCs and iPSCs exhibit an intrinsic proteostasis network that determines their self-renewal, stress tolerance, and differentiation abilities. Thus, a further characterization of this network can have important implications for extending the potential of pluripotent stem cells in cell therapy. A further step will be to examine the interconnectedness between proteostasis and the other regulators of pluripotency. For instance, it will be fascinating to define how epigenetic, transcriptional, and post-transcriptional regulatory mechanisms rewire the proteostasis network depending on the differentiation state. Likewise, in-depth analysis of how the proteostasis network impinges upon other pluripotency and differentiation factors will contribute to the understanding of ESC identity and organismal development.
Pluripotent stem cells represent an invaluable resource to generate terminally differentiated cells (e.g., neurons) facilitating the study of human diseases and drug screening. This is particularly interesting in the context of proteostasis-related diseases. A large number of human diseases result from defects in proteostasis [5], including many metabolic, cardiovascular, oncological, and neurodegenerative disorders [7]. At least 30 different human diseases are directly associated with protein misfolding and aggregation [152] and above 50 human diseases are linked to amyloid formation [5]. Whereas a collapse in proteostasis underlies these diseases, pluripotent stem cells exhibit a striking ability to correct proteostatic deficiencies. In this regard, it is important to note the remarkable ability of pluripotent stem cells to suppress the accumulation of protein aggregates [6]. For instance, iPSCs from Huntington’s disease patients do not accumulate polyQ aggregates even when they express significant amounts of mutant HTT [42, 153–155]. However, a collapse in their proteostasis network results in the accumulation of polyQ aggregates [42, 154]. Likewise, iPSCs from Machado–Joseph disease patients do not accumulate aggregates of polyQ-expanded ataxin-3 [156]. Notably, neurons derived from Huntington’s disease–iPSCs do not exhibit detectable amounts of polyQ-expanded aggregates at 12 weeks after transplantation into the brain of rat models, whereas these inclusions could be observed after 33 weeks of transplantation [154]. These observations indicate a rejuvenation process during the reprogramming process that prevents polyQ aggregation in differentiated neurons until they undergo a decline in proteostasis during the aging process. Thus, investigating pluripotent stem cells from patients could contribute to identify super-vigilant proteostasis mechanisms that can be mimicked in somatic tissues to ameliorate disease. Conversely, a demise in proteostasis of pluripotent stem cells at early developmental stages may hasten the onset of proteostasis-related diseases. In this hypothetical paradigm, pluripotent stem cells would accumulate abnormal amounts of misfolded and aggregated proteins that can be inherited by their daughter progenitor/differentiated cells. As a theoretical example, this may result in the generation of newborn neurons with a compromised proteome and polyQ-expanded HTT aggregates that could hasten Huntington’s disease-related neurodegeneration.
On the other hand, immortal pluripotent stem cells and cancer cells share many features such as high proliferation rates. Remarkably, both pluripotent stem cells and cancer cells rely on increased proteostasis mechanisms such as high expression of distinct chaperones [6, 46, 48, 157–160]. For instance, cancer cells have increased levels of HSP90s, whereas inhibitors of these chaperones could be a potential strategy for anticancer therapies [46, 160]. Moreover, the levels of CCT8 are up-regulated in hepatocellular carcinoma and gliomas, whereas its loss induces a decreased cancer proliferation and invasion [158, 159]. Proteasome activity is also increased in cancer cells, a process linked to the intrinsic requirements of these cells such as elimination of toxic proteins generated by their high mutation and proliferation rates [48]. In fact, proteasome inhibition treatments are considered a potential therapeutic approach for cancer [48]. Likewise, increased autophagic flux is involved in metabolic homeostasis and cell viability of cancer stem cells, ultimately leading to resistance to hypoxia, starvation and anticancer treatment [161]. Thus, a further understanding of increased proteostasis in pluripotent stem cells could also have important implications for cancer research and therapies. For instance, defining how the proteostasis network impinges upon immortality of pluripotent stem cells could lead to a better understanding of how cancer stem cells generate in an organism, an essential step to find specific treatments against these cells. Theoretically, alterations in proteostasis at the pluripotent stem cell level could also be a risk factor for cancer. For instance, dysregulation of enhanced proteostasis of pluripotent stem cells may result in high oxidative stress and concomitant accumulation of genotoxic factors [6] as well as misfolding of tumour suppressor factors such as p53 [162]. Conversely, a failure in the down-regulation of proteostasis mechanisms (e.g., HSP90s, TRiC/CCT assembly, and proteasome activity) during differentiation could result in stress-resistant differentiated cells with enhanced proteostasis and, eventually, higher risk of becoming tumorigenic cells. Altogether, the study of proteostasis of pluripotent stem cells could contribute to multiple fields of research such as aging, neurodegenerative disorders, and cancer.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (VI742/1-1 and CECAD).
Abbreviations
- CMA
Chaperone-mediated autophagy
- ER
Endoplasmic reticulum
- HSPs
Heat-shock chaperone proteins
- HSR
Heat-shock response
- hESCs
Human embryonic stem cells
- iPSCs
Induced pluripotent stem cells
- JDPs
J-domain proteins
- mESCs
Mouse embryonic stem cells
- NSCs
Neural stem cells
- NEFs
Nucleotide exchange factors
- UPS
Ubiquitin proteasome system
- UPR
Unfolded protein response
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353(6294):aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU. Cellular homeostasis and aging. Annu Rev Biochem. 2016;85:1–4. doi: 10.1146/annurev-biochem-011116-110806. [DOI] [PubMed] [Google Scholar]
- 3.Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 4.Powers ET, et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 5.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HJ, Gutierrez-Garcia R, Vilchez D. Embryonic stem cells: a novel paradigm to study proteostasis? FEBS J. 2017;284(3):391–398. doi: 10.1111/febs.13810. [DOI] [PubMed] [Google Scholar]
- 7.Vilchez D, Saez I, Dillin A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun. 2014;5:5659. doi: 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Prat L, Sousa-Victor P, Munoz-Canoves P. Proteostatic and metabolic control of stemness. Cell Stem Cell. 2017;20(5):593–608. doi: 10.1016/j.stem.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Vilchez D, Simic MS, Dillin A. Proteostasis and aging of stem cells. Trends Cell Biol. 2014;24(3):161–170. doi: 10.1016/j.tcb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Koyuncu S, et al. Defining the general principles of stem cell aging: lessons from organismal models. Curr Stem Cell Rep. 2015;1(3):162–169. doi: 10.1007/s40778-015-0017-1. [DOI] [Google Scholar]
- 11.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9(10):759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3(5):a004440. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberger L, et al. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17(3):155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Miura T, Mattson MP, Rao MS. Cellular lifespan and senescence signaling in embryonic stem cells. Aging Cell. 2004;3(6):333–343. doi: 10.1111/j.1474-9728.2004.00134.x. [DOI] [PubMed] [Google Scholar]
- 17.Aguilo F, et al. Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015;17(6):689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15(3):271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You KT, Park J, Kim VN. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev. 2015;29(19):2004–2009. doi: 10.1101/gad.267112.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4(10):a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavitt GD, Ron D. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb Perspect Biol. 2012;4(6):a012278. doi: 10.1101/cshperspect.a012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buszczak M, Signer RA, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014;159(2):242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Signer RA, et al. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509(7498):49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fichelson P, et al. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol. 2009;11(6):685–693. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
- 25.Insco ML, et al. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell. 2012;11(5):689–700. doi: 10.1016/j.stem.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson B, Wickens M, Kimble J. Translational control in development. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 507–544. [Google Scholar]
- 27.Zhang Q, Shalaby NA, Buszczak M. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science. 2014;343(6168):298–301. doi: 10.1126/science.1246384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savic N, et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 2014;15(6):720–734. doi: 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Sampath P, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9(8):e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29(2):108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 36.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016;19(1):66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 39.Brehme M, et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014;9(3):1135–1150. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131(1):121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Albanese V, et al. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124(1):75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 42.Noormohammadi A, et al. Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat Commun. 2016;7:13649. doi: 10.1038/ncomms13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prinsloo E, et al. Chaperoning stem cells: a role for heat shock proteins in the modulation of stem cell self-renewal and differentiation? BioEssays. 2009;31(4):370–377. doi: 10.1002/bies.200800158. [DOI] [PubMed] [Google Scholar]
- 44.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 45.Trepel J, et al. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 47.Sreedhar AS, et al. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562(1–3):11–15. doi: 10.1016/S0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 48.Saez I, Vilchez D. The mechanistic links between proteasome activity, aging and age-related diseases. Curr Genom. 2014;15(1):38–51. doi: 10.2174/138920291501140306113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradley E, et al. Regulation of embryonic stem cell pluripotency by heat shock protein 90. Stem Cells. 2012;30(8):1624–1633. doi: 10.1002/stem.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longshaw VM, et al. Knockdown of the co-chaperone Hop promotes extranuclear accumulation of Stat3 in mouse embryonic stem cells. Eur J Cell Biol. 2009;88(3):153–166. doi: 10.1016/j.ejcb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Setati MM, et al. Leukemia inhibitory factor promotes Hsp90 association with STAT3 in mouse embryonic stem cells. IUBMB Life. 2010;62(1):61–66. doi: 10.1002/iub.283. [DOI] [PubMed] [Google Scholar]
- 52.Wanderling S, et al. GRP94 is essential for mesoderm induction and muscle development because it regulates insulin-like growth factor secretion. Mol Biol Cell. 2007;18(10):3764–3775. doi: 10.1091/mbc.E07-03-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goloubinoff P. Editorial: the HSP70 molecular chaperone machines. Front Mol Biosci. 2017;4:1. doi: 10.3389/fmolb.2017.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38(10):507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 56.Finka A, Sharma SK, Goloubinoff P. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front Mol Biosci. 2015;2:29. doi: 10.3389/fmolb.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gottschling DE, Nystrom T. The upsides and downsides of organelle interconnectivity. Cell. 2017;169(1):24–34. doi: 10.1016/j.cell.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh J, Lee YD, Wagers AJ. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med. 2014;20(8):870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Son YS, et al. Heat shock 70-kDa protein 8 isoform 1 is expressed on the surface of human embryonic stem cells and downregulated upon differentiation. Stem Cells. 2005;23(10):1502–1513. doi: 10.1634/stemcells.2004-0307. [DOI] [PubMed] [Google Scholar]
- 60.Saretzki G, et al. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells. 2004;22(6):962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 61.Saretzki G, et al. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26(2):455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- 62.Gurbuxani S, et al. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene. 2003;22(43):6669–6678. doi: 10.1038/sj.onc.1206794. [DOI] [PubMed] [Google Scholar]
- 63.Battersby A, et al. Comparative proteomic analysis reveals differential expression of Hsp25 following the directed differentiation of mouse embryonic stem cells. Biochim Biophys Acta. 2007;1773(2):147–156. doi: 10.1016/j.bbamcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 64.Kaula SC, et al. Inactivation of p53 and life span extension of human diploid fibroblasts by mot-2. FEBS Lett. 2000;474(2–3):159–164. doi: 10.1016/S0014-5793(00)01594-5. [DOI] [PubMed] [Google Scholar]
- 65.Kaul SC, et al. Mortalin: present and prospective. Exp Gerontol. 2002;37(10–11):1157–1164. doi: 10.1016/S0531-5565(02)00135-3. [DOI] [PubMed] [Google Scholar]
- 66.Kaul SC, et al. Overexpressed mortalin (mot-2)/mthsp70/GRP75 and hTERT cooperate to extend the in vitro lifespan of human fibroblasts. Exp Cell Res. 2003;286(1):96–101. doi: 10.1016/S0014-4827(03)00101-0. [DOI] [PubMed] [Google Scholar]
- 67.Lopez T, Dalton K, Frydman J. The mechanism and function of group II chaperonins. J Mol Biol. 2015;427(18):2919–2930. doi: 10.1016/j.jmb.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pashai N, et al. Genome-wide profiling of pluripotent cells reveals a unique molecular signature of human embryonic germ cells. PLoS One. 2012;7(6):e39088. doi: 10.1371/journal.pone.0039088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellis RJ. Chaperonins. Curr Biol. 1999;9(10):R352. doi: 10.1016/S0960-9822(99)80223-1. [DOI] [PubMed] [Google Scholar]
- 70.Levy-Rimler G, et al. The effect of nucleotides and mitochondrial chaperonin 10 on the structure and chaperone activity of mitochondrial chaperonin 60. Eur J Biochem. 2001;268(12):3465–3472. doi: 10.1046/j.1432-1327.2001.02243.x. [DOI] [PubMed] [Google Scholar]
- 71.Leitner A, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20(5):814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spiess C, et al. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14(11):598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yam AY, et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15(12):1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faustino RS, et al. Decoded calreticulin-deficient embryonic stem cell transcriptome resolves latent cardiophenotype. Stem Cells. 2010;28(7):1281–1291. doi: 10.1002/stem.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mesaeli N, et al. Calreticulin is essential for cardiac development. J Cell Biol. 1999;144(5):857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, et al. Calreticulin reveals a critical Ca(2+) checkpoint in cardiac myofibrillogenesis. J Cell Biol. 2002;158(1):103–113. doi: 10.1083/jcb.200204092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Etchells SA, et al. The cotranslational contacts between ribosome-bound nascent polypeptides and the subunits of the hetero-oligomeric chaperonin TRiC probed by photocross-linking. J Biol Chem. 2005;280(30):28118–28126. doi: 10.1074/jbc.M504110200. [DOI] [PubMed] [Google Scholar]
- 78.Priya S, Sharma SK, Goloubinoff P. Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 2013;583(13):1981–1987. doi: 10.1016/j.febslet.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 79.Kitamura A, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8(10):1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 80.Nollen EA, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2004;101(17):6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tam S, et al. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8(10):1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Finkbeiner S. Huntington’s disease. Cold Spring Harb Perspect Biol. 2011;3(6):a007476. doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sot B, et al. The chaperonin CCT inhibits assembly of alpha-synuclein amyloid fibrils by a specific, conformation-dependent interaction. Sci Rep. 2017;7:40859. doi: 10.1038/srep40859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nillegoda NB, et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524(7564):247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bakthisaran R, Tangirala R, Rao M. Ch, Small heat shock proteins: role in cellular functions and pathology. Biochim Biophys Acta. 2015;1854(4):291–319. doi: 10.1016/j.bbapap.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 86.Mehlen P, et al. hsp27 as a switch between differentiation and apoptosis in murine embryonic stem cells. J Biol Chem. 1997;272(50):31657–31665. doi: 10.1074/jbc.272.50.31657. [DOI] [PubMed] [Google Scholar]
- 87.Buckley SM, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11(6):783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu P, et al. High autophagic flux guards ESC identity through coordinating autophagy machinery gene program by FOXO1. Cell Death Differ. 2017 doi: 10.1038/cdd.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489(7415):304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 92.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 93.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 94.Strikoudis A, Guillamot M, Aifantis I. Regulation of stem cell function by protein ubiquitylation. EMBO Rep. 2014;15(4):365–382. doi: 10.1002/embr.201338373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Werner A, Manford AG, Rape M. Ubiquitin-dependent regulation of stem cell biology. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao X, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10(6):643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adams J. The proteasome: structure, function, and role in the cell. Cancer Treat Rev. 2003;29(Suppl 1):3–9. doi: 10.1016/S0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 98.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Assou S, et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genom. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA. 2012;109(1):149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vilchez D, et al. FOXO4 is necessary for neural differentiation of human embryonic stem cells. Aging Cell. 2013;12(3):518–522. doi: 10.1111/acel.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jang J, et al. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2014;32(10):2616–2625. doi: 10.1002/stem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 104.Hernebring M, et al. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci USA. 2006;103(20):7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hernebring M, et al. Removal of damaged proteins during ES cell fate specification requires the proteasome activator PA28. Sci Rep. 2013;3:1381. doi: 10.1038/srep01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14(2):70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 107.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 108.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12(9):823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rodolfo C, Di Bartolomeo S, Cecconi F. Autophagy in stem and progenitor cells. Cell Mol Life Sci. 2016;73(3):475–496. doi: 10.1007/s00018-015-2071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 111.Ravikumar B, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90(4):1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 112.Bennett EJ, et al. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17(3):351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 113.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6(4):352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 114.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2(12):a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2010;21(3):142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 118.Egan D, et al. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7(6):643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 121.Rubinsztein DC, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1(1):11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 122.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321(5885):117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 123.Rojansky R, Cha MY, Chan DC. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife. 2016;5:e17896. doi: 10.7554/eLife.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guan JL, et al. Autophagy in stem cells. Autophagy. 2013;9(6):830–849. doi: 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pan H, et al. Autophagic control of cell ‘stemness’. EMBO Mol Med. 2013;5(3):327–331. doi: 10.1002/emmm.201201999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu K, et al. ATG3-dependent autophagy mediates mitochondrial homeostasis in pluripotency acquirement and maintenance. Autophagy. 2016;12(11):2000–2008. doi: 10.1080/15548627.2016.1212786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sotthibundhu A, et al. Rapamycin regulates autophagy and cell adhesion in induced pluripotent stem cells. Stem Cell Res Ther. 2016;7(1):166. doi: 10.1186/s13287-016-0425-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tra T, et al. Autophagy in human embryonic stem cells. PLoS One. 2011;6(11):e27485. doi: 10.1371/journal.pone.0027485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jang J, et al. Primary cilium-autophagy-Nrf2 (PAN) axis activation commits human embryonic stem cells to a neuroectoderm fate. Cell. 2016;165(2):410–420. doi: 10.1016/j.cell.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 130.Ma T, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol. 2015;17(11):1379–1387. doi: 10.1038/ncb3256. [DOI] [PubMed] [Google Scholar]
- 131.Wang S, et al. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13(5):617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 132.Wu Y, et al. Autophagy and mTORC1 regulate the stochastic phase of somatic cell reprogramming. Nat Cell Biol. 2015;17(6):715–725. doi: 10.1038/ncb3172. [DOI] [PubMed] [Google Scholar]
- 133.Ogrodnik M, et al. Dynamic JUNQ inclusion bodies are asymmetrically inherited in mammalian cell lines through the asymmetric partitioning of vimentin. Proc Natl Acad Sci USA. 2014;111(22):8049–8054. doi: 10.1073/pnas.1324035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rujano MA, et al. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4(12):e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saarikangas J, Barral Y. Protein aggregates are associated with replicative aging without compromising protein quality control. Elife. 2015;4:1–24. doi: 10.7554/eLife.06197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moore DL, et al. A mechanism for the segregation of age in mammalian neural stem cells. Science. 2015;349(6254):1334–1338. doi: 10.1126/science.aac9868. [DOI] [PubMed] [Google Scholar]
- 137.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 138.Yang J, et al. Neural differentiation and the attenuated heat shock response. Brain Res. 2008;1203:39–50. doi: 10.1016/j.brainres.2008.01.082. [DOI] [PubMed] [Google Scholar]
- 139.Araki K, Nagata K. Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol. 2011;3(11):a007526. doi: 10.1101/cshperspect.a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 141.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 142.Cho YM, et al. Induction of unfolded protein response during neuronal induction of rat bone marrow stromal cells and mouse embryonic stem cells. Exp Mol Med. 2009;41(6):440–452. doi: 10.3858/emm.2009.41.6.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu H, et al. Unfolded protein response is required for the definitive endodermal specification of mouse embryonic stem cells via Smad2 and beta-catenin signaling. J Biol Chem. 2014;289(38):26290–26301. doi: 10.1074/jbc.M114.572560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Iwawaki T, et al. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci USA. 2009;106(39):16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kratochvilova K, et al. The role of the endoplasmic reticulum stress in stemness, pluripotency and development. Eur J Cell Biol. 2016;95(3–5):115–123. doi: 10.1016/j.ejcb.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 146.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 147.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 148.Bertolotti A, et al. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 149.Shen J, et al. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 150.Luo S, et al. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol Cell Biol. 2006;26(15):5688–5697. doi: 10.1128/MCB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol. 2011;3(7):a007559. doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cuanalo-Contreras K, Mukherjee A, Soto C. Role of protein misfolding and proteostasis deficiency in protein misfolding diseases and aging. Int J Cell Biol. 2013;2013:638083. doi: 10.1155/2013/638083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11(2):264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jeon I, et al. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30(9):2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 155.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Koch P, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado–Joseph disease. Nature. 2011;480(7378):543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 157.Boudiaf-Benmammar C, Cresteil T, Melki R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS One. 2013;8(4):e60895. doi: 10.1371/journal.pone.0060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Huang X, et al. Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS. 2014;122(11):1070–1079. doi: 10.1111/apm.12258. [DOI] [PubMed] [Google Scholar]
- 159.Qiu X, et al. Overexpression of CCT8 and its significance for tumor cell proliferation, migration and invasion in glioma. Pathol Res Pract. 2015;211(10):717–725. doi: 10.1016/j.prp.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 160.Rappa F, et al. HSP-molecular chaperones in cancer biogenesis and tumor therapy: an overview. Anticancer Res. 2012;32(12):5139–5150. [PubMed] [Google Scholar]
- 161.Lei Y, et al. Targeting autophagy in cancer stem cells as an anticancer therapy. Cancer Lett. 2017;393:33–39. doi: 10.1016/j.canlet.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 162.Trinidad AG, et al. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50(6):805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]