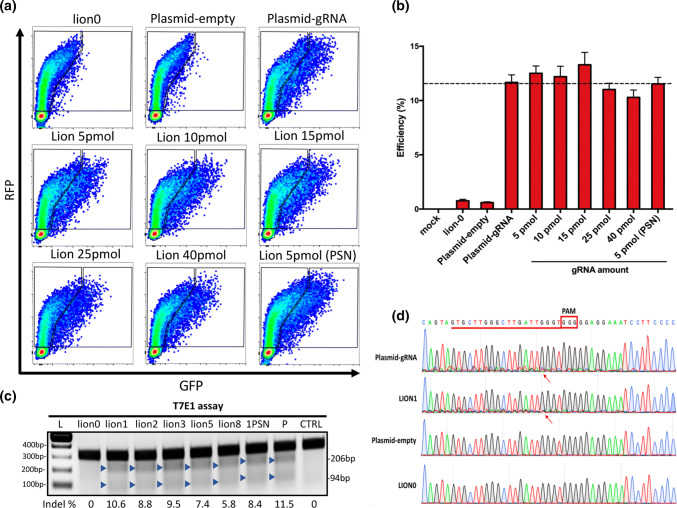
Fig. 3.
Optimization of gRNA amounts used in LION and validation of LION CRISPR vector-based gene editing. a FCM patterns of C-Check efficiency tested for different amounts of gRNA used in LION CRISPR vector generation. b FCM quantification of C-Check cleavage efficiency in a comparison between traditional and LION CRISPR vectors (n = 3). c T7E1 assay results of LION and pure CRISPR plasmid. L indicates 100 bp DNA ladders. Lion 0, 1, 2, 5, 8 indicate 1000 ng backbone ligated with 0, 1, 2, 3, 5, 8 μL annealed gRNA (5 μM), respectively. 1PSN represents treatment of LION CRISPR made from 5 pmol annealed gRNA oligo and treated with plasmid-safe DNA nuclease. P indicates pure plasmid. CTRL indicates negative control. d Sanger sequencing to detect indel spectrums of optimized LION and pure plasmids. Red line and frame indicate the gRNA sequence. Red arrows indicate the cleavage site of CRISPR/Cas9