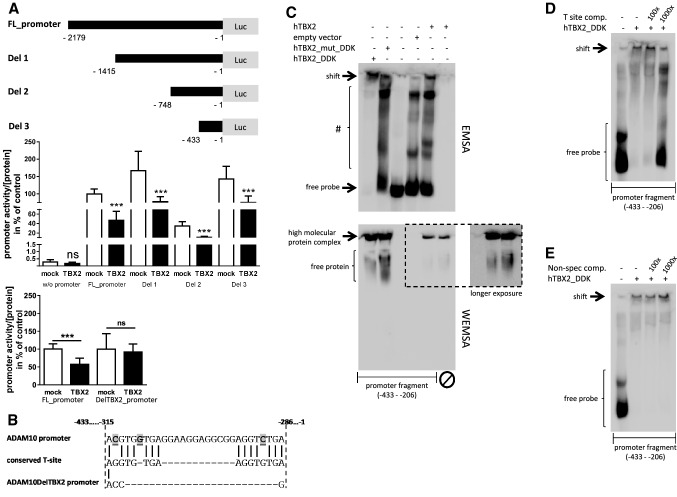
Fig. 4.
Identification of TBX2 binding sites in the human ADAM10-promoter. a Schematic overview of ADAM10-promoter constructs with sequential sequence deletion. Effect of TBX2 overexpression on ADAM10 deletion mutants and on a mutant lacking the TBX2 binding sites (DelTBX2_promoter) was analyzed in comparison to the full-length promoter sequence (FL_promoter) by luciferase-based reporter gene assay. Values represent mean ± SD from three independent experiments performed in triplicate. (ns p > 0.05, ***p < 0.001, Unpaired Student’s t test). b A putative TBX2 binding sequence within the human ADAM10-promoter (− 315 to − 286 bp upstream of the translation initiation site) consisting of two partially conserved T-sites. Sequence discrepancy between the ADAM10-promoter and consensus T-site sequence is highlighted. Additionally, the sequence of the mutant lacking the binding sites is shown. c Electrophoretic mobility shift assay (EMSA) for the identified region of the ADAM10-promoter sequence comprising a putative TBX2 binding motif. The biotinylated promoter fragment (− 433 to − 206) was incubated with nuclear fractions from HEK 293 cells either overexpressing DDK-tagged TBX2, DDK-tagged mutant TBX2 (G121A/R122S), untagged TBX2 or fractions from empty vector transfected cells. Protein-DNA complexes were separated from the free DNA probe by electrophoresis in a native polyacrylamide gel and chemiluminescent signals were visualized by HRP-coupled streptavidin conjugate. Free probe and high molecular weight, TBX2 containing complexes (indicated as shift) are marked. Formed DNA–protein complexes with lower molecular weight are indicated with #. In addition, TBX2 proteins were visualized by western blot-EMSA (WEMSA) using a TBX2-specific antibody under the same conditions as for EMSA analysis. d Nuclear extracts from human TBX2_DDK expressing cells were incubated with the promoter fragment and separated as described in c with or without the presence of an unlabeled competitive oligonucleotide comprising a fully conserved palindromic T-site ([54], see “Methods” section) or an unrelated oligonucleotide, lacking any binding potency towards TBX2, as a specificity control (e). Molar excess of unlabeled positive oligonucleotide is indicated