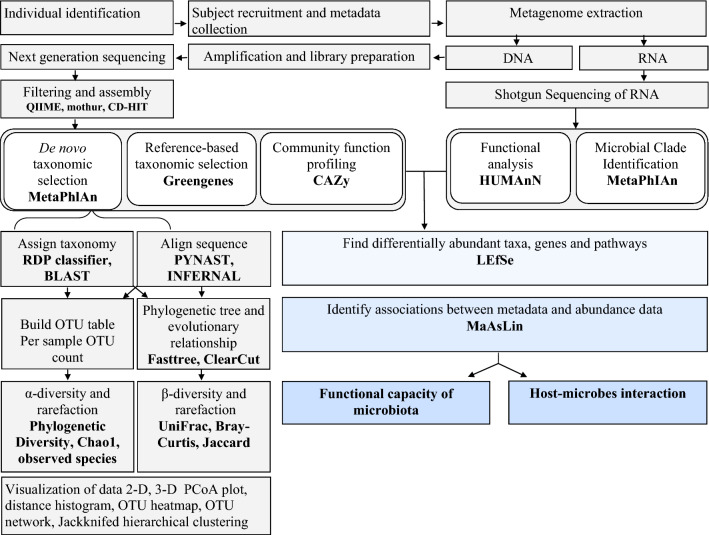
Fig. 1.
Bioinformatics work flow of culture independent approach. QIIME quantitative insights into microbial ecology, MG-RAST metagenomics rapid annotation using subsystem technology, CAZy carbohydrate active-enzymes, MetaPhlAn metagenomic phylogenetic analysis, KEGG Kyoto encyclopaedia for genes and genomics, COG clusters of orthologous group, PICRUst phylogenetic investigation of communities by reconstruction of unobserved states, HUMAnN The Human Microbiome Project Unified Metabolic Analysis Network, LEfSe Linear Discriminate Analysis with Effect Size, MaAsLin Multivariate Association with Linear Models, MetaPhlAn Metagenomic Phylogenetic Analysis, PICRUSt Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, rRNA ribosomal RNA