Table 3.
Description of FUBP1-inhibitors (top of the list) and activator (last in the list) identified so far
| Compound | Chemical structure | Mechanism | Effect on FUBP1 target genes | IC50 |
|---|---|---|---|---|
| Benzoyl anthranilic Acid [150] |
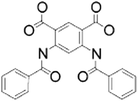
|
Interacts with the hydrophobic region of the KH domain of FUBP1, leading to an impaired DNA-binding | ↓ Binding on MYC | 350 μM |
| Pyrazolo[1,5a] pyrimidine [11] |
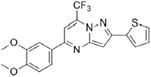
|
Inhibits or prevents the binding between FUBP1 and the FUSE |
↓ Binding on P21 and ↑ P21 mRNA ↓ Binding on BIK and ↑ BIK mRNA ↓ CCND mRNA in Hep3B cells |
11–24 μM depending on assays |
| GSK343 [128] |
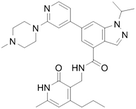
|
Competitive inhibitor | ↓ MYC protein level in Saos-2 cells | Not available (4 nM for EZH2) |
| Camptothecin (CPT) [156] |
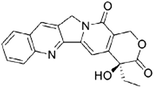
|
Interferes with the transcriptional control of FUBP1 |
↑ P21 mRNA ↑ BIK mRNA ↓ CCND2 mRNA ↓ TCTP mRNA in Hep3B and HepG2 cells. |
3.2 ± 0.6 mM |
| 7-ethyl-10-hydroxycamptothecin (SN-38) [156] |
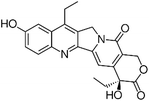
|
Interferes with the transcriptional control of FUBP1 |
↑ P21 mRNA ↑ BIK mRNA ↓ CCND2 mRNA ↓ TCTP mRNA in Hep3B and HepG2 cells. |
0.78 ± 0.2 mM - 1.9 ± 0.7 mM depending on assays |
| ACTIVATOR: pyrido-pyrimidinone SMN-C2 and SMN-C3 [144] |
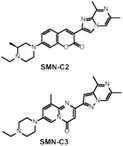
|
Binds to the AGGAAG motif on exon 7 of the SMN2 pre-mRNA Creates a new functional binding surface with FUBP1 |
↑ SMN2 splicing | EC50: 100 nM |
