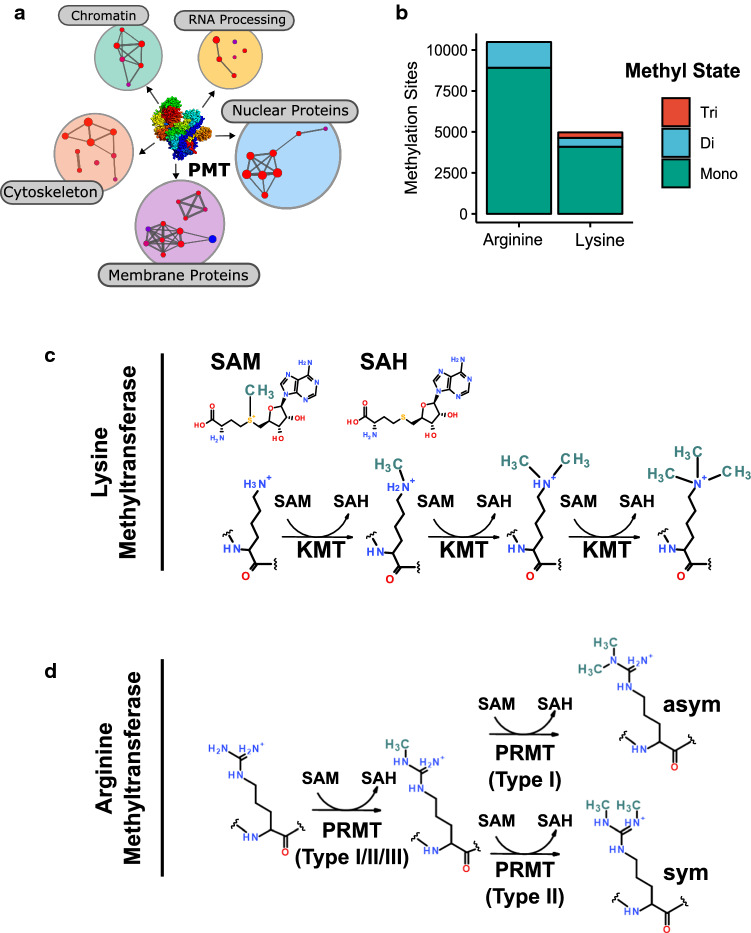
Fig. 1.
Protein methylation in humans. a Protein methyltransferase substrates are found throughout the cell. Shown is a network view of associated cellular component gene ontologies of methyltransferase substrates identified in the PhosphoSite database [8]. b Number of arginine and lysine methylation sites in the phosphosite database by methyl state. c, d Schematic illustrations of lysine and arginine methylation. Lysine can be mono-, di-, or trimethylated by KMT enzymes at the ε-amino group. Type I/II/III PRMTs can monomethylate arginine on guanidine group. Further modification by Type I PRMTs results in asymmetric dimethylation, while Type II proceeds to symmetrical dimethylation of arginine residues