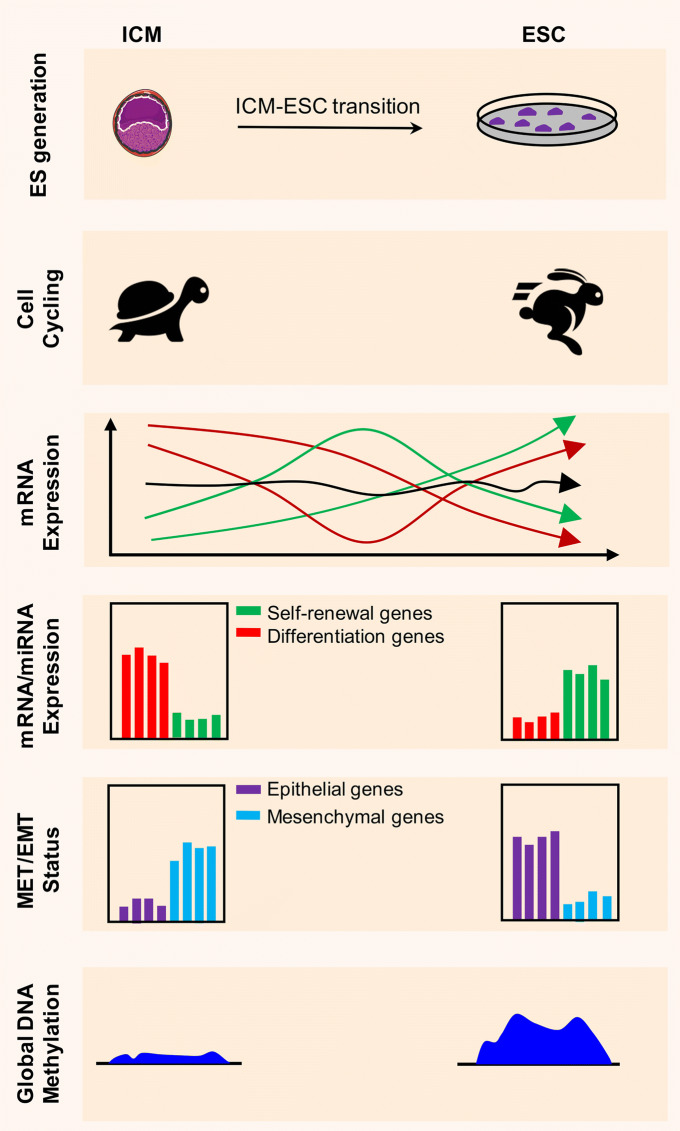
Fig. 5.
Most notable mechanistic events occurring during ICM-ESC transition. ESCs are generated from the ICM-derived EPI cells through a process which appears to be a combination of reprogramming and capturing. Although ESCs exhibit a transcriptome highly similar to EPI cells, they are highly proliferative as opposed to ICM/EPI cells which are almost in a silent condition in terms of cell cycling. Genes are highly differentially expressed over the course of ESC formation. Some transcripts are either upregulated (green lines in ‘mRNA Expression’ part) or downregulated (red lines in ‘mRNA Expression’ part), whereas others remain unchanged during ICM outgrowth (black line in ‘mRNA Expression’ part). Upregulated transcripts such as, Sox2, Nr0b1, and Id1 appear to be necessary for ESC formation, while downregulated ones such as Cdx2, Tgfbr2, and Gata6 appear to have adverse effects on ICM transition to ESCs. Protein-coding (e.g. Nanog and Eras) and non-coding genes (e.g. miR-302 miRNAs) associated with self-renewal are highly induced, while those associated with differentiation (e.g. Sox17, Hoxd8, and some of let-7 miRNAs) exhibit downregulation. Importantly, epithelial-associated mRNAs and miRNAs show increased expression while those affiliated with mesenchymal phenotype are downregulated during ICM outgrowth into ESCs. Finally, in contrast to ICM cells which exhibit a global DNA hypomethylation, ESCs grown in culture media other than 2i/L mostly show genome-wide hypermethylation of DNA