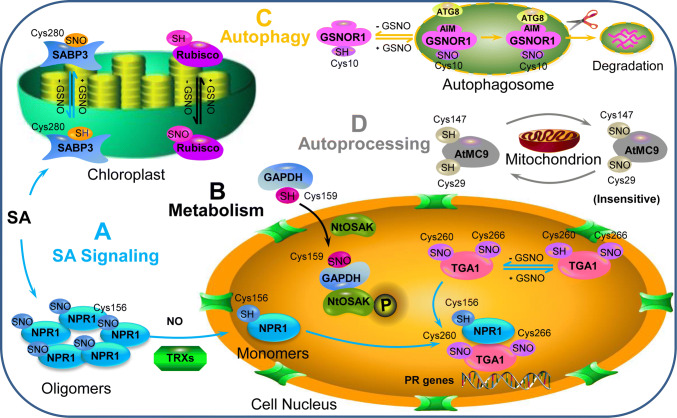
Fig. 2.
S-nitrosylation of SA signaling-related and the metabolism- and photosynthesis -related proteins GSNOR and MC9 involved in PCD regulation. aS-nitrosylation regulates SA signaling (lines in blue): SABP3 can be S-nitrosylated at its Cys-280 during the SA-related defense response. Under normal conditions, S-nitrosylation is beneficial for NPR1 in an oligomeric state in the cytosol. The oligomer can be reduced to monomers by TRXs in a NO-dependent manner and then translocated to the nucleus; S-nitrosylation of TGA1 at two Cys sites results in the formation of the NPR1/TGA system, regulating the expression of downstream PR genes. bS-nitrosylation of two metabolism-related proteins (lines in black): Rubisco and GAPDH can be S-nitrosylated, and S-nitrosylated GAPDH can interact with NtOSAK to respond to stress. c GSNOR S-nitrosylation leads to GSNOR degradation via selective autophagy (lines in orange); dS-nitrosylation of MC9 (lines in gray). SA salicylic acid, SABP3 salicylic acid-binding protein 3, GSNO S-nitrosoglutathione, NPR1 nonexpresser of pathogenesis-related Genes 1, NO nitric oxide, TRXs thioredoxins, TGA1 TGACG motif binding factor1, Rubisco ribulose-1,5-bisphosphate carboxylase/oxygenase, GAPDH glyceraldehyde 3-phosphate dehydrogenase, NtOSAK Nicotiana tabacum osmotic stress-activated protein kinase, GSNOR1 S-nitrosoglutathione reductase 1, ATG8 AUTOPHAGY-RELATED8, AIM ATG8-interacting motif, AtMC9 A. thaliana metacaspase 9