Abstract
Lung cancer remains the leading cause of cancer-related death worldwide, and the high incidence rates are worrisome. Exosomes are a class of extracellular vesicles secreted by most cells, including RNAs, proteins and lipids. Exosomes can mediate cell-to-cell communication in both physiologic and pathologic processes. Accumulated evidences show that cancer-derived exosomes aid in the recruitment and reprogramming of constituents correlated with tumor microenvironment. Furthermore, exosome-based clinical trials have been completed in advanced lung cancer patients. In this review, we discuss the roles of exosomes in a lung cancer microenvironment, such as its participation in lung cancer initiation, progression and metastasis as well as being involved in angiogenesis, epithelial–mesenchymal transition (EMT), immune escape, and drug resistance. In addition, we focus on the potential of exosomes as diagnostic and prognostic biomarkers in lung cancer, as well as the challenges faced by and advantages of exosomes as drug delivery vehicles and in exosome-based immunotherapy.
Keywords: Lung cancer, Exosomes, Biomarkers, Immunotherapy, Drug delivery vehicles
Introduction
Lung cancer remains the leading cause of cancer-related death worldwide with 1.8 million deaths predicted in 2018, accounting for almost 1 in 5 (18.4%) cancer deaths [1]. Lung cancer is divided into small-cell lung cancer (SCLC, about 15%) and non-small-cell lung cancer (NSCLC, about 85%) containing adenocarcinoma and squamous cell carcinoma [2]. Of importance, great progress has been made in the treatment of NSCLC, such as epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) [3–7], anaplastic lymphoma kinase (ALK) inhibitors [8], and bevacizumab [9], as well as immune checkpoints inhibitors [10–17]. However, only a small proportion of patients benefit from these drugs. In recent years, the biofunctional and biochemical nature of exosomes have been extensively studied, which can shed new light on the molecule mechanism of lung cancer initiation and progression, and contribute to developing new diagnostic and therapeutic strategies for lung cancer patients.
Exosomes are a class of extracellular vesicles (EVs) of endocytic origin with a diameter of 30–100 nm and a density in sucrose gradients of 1.13–1.19 g/ml [18–20]. Vesicles released during the maturation of sheep reticulocytes were first defined as exosomes by Johnstone et al. [21].
Exosomes are membrane-bound phospholipid vesicles, including proteins, nucleic acids (mRNA and noncoding RNAs) and lipids [19, 22]. Exosomes are secreted by most cell types, in particular tumor cells; tumor-derived exosomes play an important role in the communication between tumor cells and their microenvironment, contributing to creating a favorable soil for tumor progression [23]. Noteworthy, exosomes have been isolated in most biological fluids, such as serum and plasma, saliva, urine, breast milk, semen, amniotic fluid, cerebrospinal fluid, pleural effusions, bronchial lavage, ascites fluid, synovial fluid and bile [24–27]. Moreover, exosomes provided a protective vesicle for transported small RNAs against degradation of RNase [28]. These special features make exosome an ideal specimen for liquid biopsy. In addition, as the therapeutic potential of exosome was explored, exosome-based phase I and II clinical trials have been completed in advanced lung cancer patients, with a favorable safety profile [29, 30]. On the other hand, exosomes have recently come into focus as drug delivery vehicles that can deliver chemotherapy drugs and biologics for lung cancer therapy [31–33].
Here, we describe how exosomes are formed and released into extracellular space, and discuss the advantages and limitations of some techniques used for exosomes isolation and purification. We also summarize the most recent studies on exosomes that participated in the reprogramming of the lung cancer microenvironment. Lastly, we focus on the roles of exosomes as potential biomarkers and drug delivery vehicles, as well as in exosome-based immunotherapy.
Formation, release and uptake of exosomes
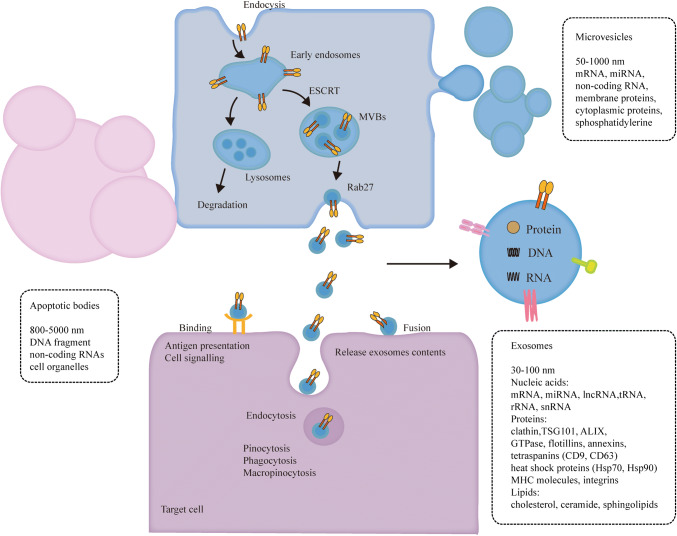
Extracellular vesicles are a heterogeneous population of membrane vesicles that based on their biogenesis are classified into three distinct classes: exosomes, microvesicles and apoptotic bodies (Fig. 1) [34–39]. Exosome is initiated when membrane proteins are endocytosed via inward budding of the plasma membrane to form the early endosome, then, early endosomes mature into late endosomes, which are also known as multivesicular bodies (MVBs) containing numerous intraluminal vesicles (ILVs) formed by the invagination of the endosomal membrane [35]. During invagination, cytosolic and membrane proteins, lipids, and RNAs are incorporated into the ILVs [40, 41]. MVBs can fuse with lysosomes, resulting in the degradation of cargos carried by vesicles [40, 42]. Alternatively, MVBs may fuse with the plasma membrane, releasing exosomes to the extracellular space [43].
Fig. 1.
Formation, release and uptake of exosomes. Extracellular vesicles are classified into three distinct classes: exosomes, microvesicles and apoptotic bodies. Apoptotic bodies are released from the plasma membrane during cell undergoing programmed cell death [32]. Microvesicles are shed directly outwards budding from the plasma membrane [33, 34]. Exosome formation starts when membrane proteins are endocytosed via inward budding of the plasma membrane to form the early endosome and exosomes are formed as intraluminal vesicles (ILVs) by the invagination of the endosomal membrane. Finally, exosomes are released by fusion of the MVBs with the plasma membrane. Several machineries are involved in sorting of cargos into ILVs and formation of ILVs. When exosomes bind to the recipient cells, they may remain bound to the surface and induce intracellular signaling, or they directly fuse with the plasma membrane and release their contents into the cytoplasm. Additionally, exosomes can be internalized by clathrin-independent or clathrin-dependent endocytosis, including pinocytosis, phagocytosis and macropinocytosis
The endosomal sorting complex required for transport (ESCRT) and associated proteins (Clathrin, TSG101 and ALIX) are involved in ubiquitinated proteins sorting and ILVs formation [44–49]. However, the cargos sorting and ILVs formation are not solely dependent on ESCRT complexes, but required the sphingolipid ceramide [50]. In addition, tetraspanins are also involved in the biogenesis of exosomes, such as CD9, CD63, CD81 and CD82 enriched in exosomes from various types of cells, which are able to form oligomeric complexes and interact with integrins and MHC molecules [51, 52]. The Rab family of small GTPases, for example, Rab5 is required for the biogenesis of the early endosomes, late endosomes and lysosomes [53, 54], Rab7 regulate the transfer of cargos from the MVBs to the lysosomes [55], Rab35, Rab27a and Rab27b are involved in MVBs fusion with plasma membrane and exosomes secretion [56, 57]. However, the mechanisms that control the sorting of specific RNA species to exosomes are yet to be shown. Kosaka et al. showed that miRNAs can be loaded into exosomes and released through a ceramide-dependent pathway but not ESCRT machinery [58]. Villarroya-Beltri et al. reported that the protein heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) controlled miRNAs sorting to exosomes through binding to a specific motif of miRNA [59]. Squadrito et al. demonstrated that miRNA loading into exosomes was regulated by cell activation-dependent changes in miRNA target levels in the parental cells [60].
Owing to their endosomal origin, nearly all exosomes contain proteins involved in MVBs biogenesis (clathrin, TSG101 and ALIX), intracellular membrane transport and fusion (GTPases, flotillins and annexins), and tetraspanins (CD9 and CD63), which are used as surface markers for exosomes identification [61]. Exosomes contain heat shock proteins (Hsp70 and Hsp90) involved in antigen presentation and package peptides into MHC molecules [62]. Exosomes also contain integrins that possibly address exosomes to recipient cells [18]. In addition, exosomes are enriched in lipid rafts, such as cholesterol, ceramide and sphingolipids, which show difference in the distribution of membrane lipids from their parental cells [63].
Then, exosomes get to the recipient cells, after that they may bind to the surface receptors and induce intracellular signaling [64, 65]. Exosomes can be taken up by recipient cells through clathrin-dependent and clathrin-independent endocytosis, such as pinocytosis, phagocytosis and macropinocytosis [66–69]. Additionally, exosomes also can be internalized by caveolae and lipid raft-mediated endocytosis [41]. On the other hand, exosomes could directly fuse with the plasma membrane and release their contents into the cytoplasm [70].
Isolation and purification of exosomes
Exosomes isolation and purification from cell culture supernatants and biological fluids are essential to biomedical investigation and clinical application. In general, the isolation methods of exosomes are mainly based on the physical (such as size, density and molecular weight), chemical (surface charge) and biological (specific markers) properties of exosomes. Ultracentrifugation is the most widely used conventional protocol for exosomes isolation, which involves a series of centrifugations at increasing speed to remove dead cells and large apoptotic debris, followed by a final high-speed ultracentrifugation (100,000g for 70 min) to precipitate exosomes [71]. Nevertheless, the method requires a lot of time and costly instruments (ultracentrifuge), and precipitated exosomes may contain aggregated proteins [72]. Owing to exosomes have a floatation density of 1.13–1.19 g/ml on sucrose gradients [18], based on extracellular vesicles different floatation densities, using sucrose-gradient centrifugation helps to further separate different vesicular density [73–75], therefore, this method is thought to precipitate exosomes at a higher purity. Recently, several commercial kits, for example, ExoQuick™ (System Biosciences) that rely on polymer co-precipitation have been widely used to isolate exosomes [76]. The precipitated exosomes via ExoQuick™ could be easily and quickly isolated with low centrifugal forces, whereas this approach is likely to co-isolate heterogeneous polymeric particles and lack specificity [77]. Exosomes express abundant membrane proteins, such as MHC I and II molecules, costimulatory molecules, and adhesion molecules that can be used as markers for exosomes immune isolation. Clayton et al. utilized magnetic beads, coated with monoclonal antibodies specific to HLA DR, DQ and DP for the particular isolation of exosomes from cell-free supernatants, which is a rapid protocol for the isolation of antigen-presenting cells (APCs) exosomes [78]. In particular, ultrafiltration is suitable for purifying exosomes from large volumes (> 1 L) of conditioned medium [71]. Lamparski et al. combined ultrafiltration and ultracentrifugation for the purification of clinical-grade exosomes derived from APCs [79]. On the other hand, size-exclusion purification is comparable to density gradient isolation of exosomes [80]. Boing et al. used size-exclusion chromatography (SEC) to isolate extracellular vesicles from human plasma and they investigated that SEC purified extracellular vesicles with diameter larger than 70 nm from human platelet-free supernatant of platelet concentrates, moreover, the vesicle pellets less than 5% of HDL and less than 1% of protein [81]. In addition, field flow fractionation [82] and microfluidic chip [83] are also developed to elevate purification efficiency. Of note, fetal bovine serum (FBS) also contains exosomes and influences cultured cell phenotypes, thus, conditioned medium was replaced by FBS eliminated of exosomes via 18 h ultracentrifugation before the exosomes were extracted [84]. After isolation and purification of exosomes from cell culture supernatant or other biological fluids, the vesicles size distribution, morphologies and surface markers were further analyzed to verify that they are exosomes, and the isolation and detection technologies of exosomes are summarized in Table 1.
Table 1.
Isolation and detection technologies for exosomes
| Contents | Platform | References |
|---|---|---|
| Isolation and purification methods | Ultracentrifugation | [71, 72] |
| Sucrose-gradient centrifugation | [73–75] | |
| Polymer co-precipitation | [76, 77] | |
| Antibody-coated magnetic beads | [78] | |
| Ultrafiltration | [71, 79] | |
| Size-exclusion chromatography | [81] | |
| Field flow fractionation | [82] | |
| Microfluidic chip | [83] | |
| Analysis of morphology and size distribution | Transmission electron microscopy (TEM) | [72] |
| Scanning electron microscopy (SEM) | [85] | |
| Cryo-electron microscopy (cryo-EM) | [86] | |
| Atomic force microscopy (AFM) | [87] | |
| Analysis of concentration and size distribution | Nanoparticle tracking analysis (NTA) | [88] |
| Dynamic light scattering (DLS) | [89] | |
| Tunable resistive pulse sensing (TRPS) | [90] | |
| Protein quantification and characterization | Western blotting | [80] |
| Enzyme-linked immunosorbent assay (ELISA) | [91] | |
| Mass spectrometry | [92] | |
| Nucleic acid amplification and sequencing | Polymerase chain reaction (PCR) | [93] |
| Next-generation sequencing (NGS) | [94] |
The role of exosomes in the lung cancer microenvironment
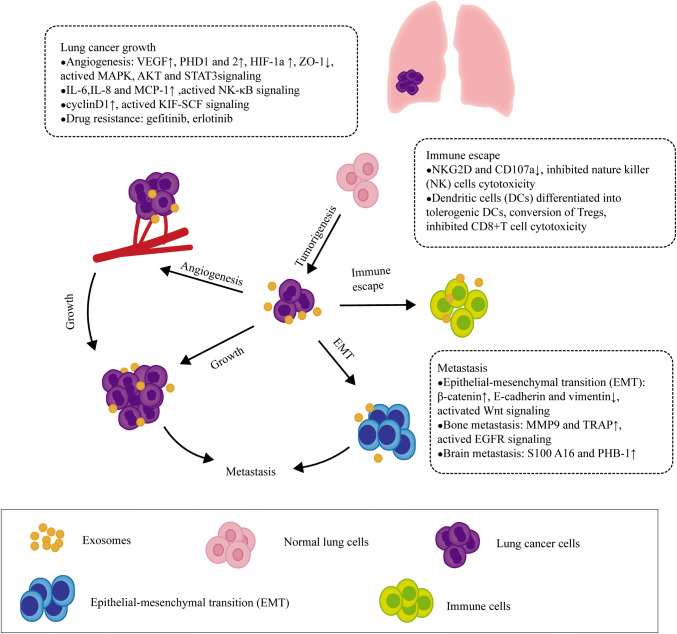
Exosomes contain RNAs, proteins, and lipids, all of which affect exosomes functions and reflect cell characteristics. The major nucleic acids of exosomes are RNAs that contain microRNA (miRNA), tRNA, and long noncoding RNA (lncRNA), as well as largely fragmented mRNAs [95, 96]. Exosomes transfer RNAs to recipient cell, mRNA may be translated into proteins, and miRNAs can modulate the translation of target mRNA in host cells [97]. In addition, exosomal proteins can be also transported to recipient cells and trigger strong cellular responses [98]. Exosomes can serve as a novel cellular communication by transferring their contents to target cells in a lung cancer microenvironment, thereby regulating lung cancer cells progression (Fig. 2 and Table 2).
Fig. 2.
The role of exosomes in a lung cancer microenvironment. Exosomes mediate communication between lung cancer cells and their surrounding microenvironment through transferring their biologically active cargos as well as participate in lung cancer initiation, progression and metastasis involved in angiogenesis, epithelial–mesenchymal transition (EMT), immune escape and drug resistance
Table 2.
Exosomal cargos detected in lung cancer and clinical relevance
| Exosomal Cargos | Parental cells | Recipient cells | Target | Biological/clinical relevance | Reference |
|---|---|---|---|---|---|
| (1) miRNA | |||||
| miR-9 | H1299 cells | Endothelial cells | SOCS5 | Promoted endothelial cell migration and tumour angiogenesis | [100] |
| miR-23a | CL1-5 cells | Endothelial cells | PHD1 PHD2 | Increased endothelial permeability and cancer cells transendothelial migration | [102] |
| miR-21 | CSE-transformed HBECs | Normal HBECs | VEGF | Facilitated angiogenesis | [103] |
| miR-210 | A549L cells | HUVECs | TIMP-1 | Increased vascularization in A549L-derived tumour xenografts | [150] |
| miR-23a | (TGF)-β-treated A549 cells | A549 cells | TCF4/β-catenin | EMT | [105] |
|
miR-21 miR-29a |
A549 cells | Immune cells | TLRs | Led to tumor growth and metastasis | [113] |
| miR-142-3p |
H1437 H2073 |
Endothelial and fibroblast cells | TGFβR1(endothelial cells) | Promoted angiogenesis in endothelial cells; facilitated the CAFs transformation in lung fibroblast cells | [115] |
| miR-223 | Platelets | A549 cells | EPB41L3 | Promoted A549 cells invasion | [123] |
| miR-21 | A549 cells | Osteoclast progenitor cells | PDCD4 | Promoted osteoclastogenesis | [127] |
| miR-21 | H827R cells | HCC827 cells | Led to gefitinib resistance in HCC827 cells | [129] | |
| miR-214 | PC-9GR cells | PC9cells | Mediated gefitinib resistance | [132] | |
| (2) Protein | |||||
| EGFR | A549 cells | Endothelial cells | VEGF | Facilitated tumor angiogenesis | [101] |
| Vimentin | PC14HM cells | HBECs | EMT | [110] | |
| HSP70 | A549 cells | MSCs | TLR2 | Induced a pro-inflammatory phenotype in MSCs | [111] |
| KIT | Mast cells | A549 cells | cyclin D1 | Accelerated the proliferation of A549 cells | [112] |
| CD41 | Platelets | A549 cells | cyclin D2 | Facilitated lung cancer cells proliferation | [114] |
| EGFR | Lung cancer cells | DCs | Suppressed tumor specific CD8 + T cells | [120] | |
| MMP3 | Adipocytes | 3LL cells | Promoted 3LL cells invasion | [125] | |
| AREG | CRL-2868 | Osteoclasts |
MMP9 TRAP |
Triggered osteolytic bone metastasis | [126] |
| S100A16 | SCLC cells | HBMECs | PHB-1 | Facilitated SCLC cells brain metastasis | [128] |
| (3) lncRNA | |||||
| lnc-MMP2-2 | (TGF)-β-treated A549 cells | A549 cells | MMP2 | Regulated vascular permeability and lung cancer invasion | [104] |
| RP11-838N2.4 |
HCC4006/R HCC827/R |
HCC4006 HCC827 |
FOXO1 | Reduced resistance to erlotinib treatment | [130] |
| lncRNA H19 |
HCC4006/R HCC827/R |
HCC4006 HCC827 |
Induced resistance to gefitinib treatment | [133] | |
SOCS5 suppressor of cytokine signaling 5, PHD1 and 2 prolyl hydroxylase 1 and 2, HUVECs human umbilical vein endothelial cells, CSE cigarette smoke extract, (MMP)2-2 lnc-matrix metalloproteinase, TIMP-1 tissue inhibitor of metalloproteinases-1, HBECs human bronchial epithelial cells, MSCs mesenchymal stem cells, TLR2 Toll-like receptor 2, Lnc-MMP-2-2 lnc-matrix metalloproteinase, TLRs Toll-like receptors, CD41 glycoprotein IIb/IIIa, CAFs cancer-associated fibroblast, DCs dendritic cells, EPB41L3 the 3′UTR of erythrocyte membrane protein band 4.1-like 3, 3LL Lewis lung cancer cells, AREG EGFR binds amphiregulin, MMP9 matrix metalloproteinases 9, TRAP tartrate-resistant acid phosphatase, HBMECs human brain microvascular endothelial cells, PHB-1 prohibitin-1, FOXO1 forkhead box protein O1
Exosomes in lung cancer angiogenesis
Tumor growth and metastasis require new blood vessels to supplement oxygen and nutrients, which activate vasculogenesis and angiogenesis, and consequently the sprouting of blood vessels from the neighboring tissues into the tumor [99]. Zhuang et al. found that tumor-derived miR-9 was transferred into endothelial cells via microvesicles and that miR-9 effectively decreased the suppressor of cytokine signaling 5 (SOCS5) level, and induced the activation of JAK–STAT pathway, ultimately promoting endothelial cell migration and tumor angiogenesis [100]. Exosomes secreted by A549 cells harboring activated EGFR can be internalized by cocultured endothelial cells, which activated MAPK and AKT signaling, and induced expression of vascular endothelial growth factor (VEGF) in endothelial cells, followed by autocrine activation of VEGF receptor-2 (VEGFR-2), facilitating tumor angiogenesis [101]. Under hypoxic conditions, miR-23a was significantly increased in exosomes from lung cancer cells, and exosomes transferred miR-23a to endothelial cells; miR-23a directly inhibited its target prolyl hydroxylase 1 and 2 (PHD1 and 2) expression, resulting in the accumulation of hypoxia-inducible factor-1 α (HIF-1 α) in endothelial cells. Consequently, exosomes derived from hypoxic lung cancer cells increased endothelial permeability and cancer cells transendothelial migration in vitro, enhanced neovascularization and tumor growth in vivo [102]. Liu et al. found that smoking induced an increase in the serum miR-21 level, and exosomes isolated from cigarette smoke extract (CSE)-transformed human bronchial epithelial (HBE) cells also showed a high level of miR-21. Transformed HBE cells delivered miR-21 into normal HBE cells by exosomes, which activated IL-6/STAT3 signaling and enhanced the expression of VEGF in recipient cells, facilitating angiogenesis in HBE cells [103]. Wu et al. screened a exosomal lncRNA and lnc-matrix metalloproteinase (MMP)2-2 was remarkably enriched in transforming growth factor (TGF)-β-treated A549 cells, and promoted MMP2 expression. Coculture experiments revealed that TGF-β-mediated exosomes, as vectors of intercellular communication, regulated vascular permeability and lung cancer invasion [104]. Due to overexpression of tissue inhibitor of metalloproteinases-1 (TIMP-1) in lung cancer cells, miR-210 was up-regulated in exosomes. These exosomes facilitated tube formation activity in human umbilical vein endothelial cells (HUVECs), which was reflected in increased vascularization in A549L-derived tumor xenografts [105]. Together, these results support that exosomes as a novel pro-angiogenic mechanism can regulate vascular remodeling in tumor microenvironment by directional transfer of the angiogenic factors between tumor cells and endothelial cells, thereby promoting tumor angiogenesis.
Exosomes and epithelial–mesenchymal transition (EMT)
Epithelial–mesenchymal transition (EMT) is a highly conserved process confirmed by the loss of cell–cell adhesion and acquisition of the mesenchymal phenotype, which is in favor of tumor cells being migratory and invasive [106]. Interestingly, recent studies indicated that exosomes participated in the EMT process, during which exosomes transferred mesenchymal-associated information between the tumor cells and the microenvironment, and regulated signal transduction in recipient cells [107, 108]. Exosomes isolated from TGF-β1-induced mesenchymal cell showed that the expression of β-catenin increased and E-cadherin as well as vimentin decreased. Meanwhile, miR-23a was significantly up-related in secreted exosomes. Further investigation showed that exosome stimulated TCF4/β-catenin transcriptional activity and activated canonical Wnt signaling in A549 cells undergoing EMT [109]. Rahman MA et al. found exosomes derived from highly metastatic lung cancer PC14HM cells that express higher vimentin than those from non-metastatic lung cancer PC14 cells. When human bronchial epithelial cells (HBECs) were treated with PC14HM exosomes, vimentin expression was significantly up-related, inducing EMT in recipient HBECs [110]. These results suggest tumor-derived exosomes to be an important mediator of EMT, thereby transforming tumor cells to a more aggressive phenotype, though more studies need to be performed.
Exosomes regulate lung cancer growth
Emerging evidence indicate that exosomes play a bimodal role in cancer and they can program the local and systemic environment to help cancer growth and dissemination [23]. A549 cell-derived exosomes were internalized by mesenchymal stem cells (MSCs), which significantly stimulated IL-6, IL-8, and MCP-1 production and recruited macrophages, promoting tumor growth. It was further detected that Hsp70 on the surface of A549 cell-derived exosomes activated NF-κB signaling through Toll-like receptor 2 (TLR2), inducing a pro-inflammatory phenotype in MSCs [111]. Xiao et al. demonstrated mast cells like HMC-1-derived exosomes contain c-KIT mRNA, and HMC-1 exosomes transferred KIT protein to A549 cells, subsequently activating KIT-SCF signaling, which increased cyclin D1 expression and accelerated the proliferation of A549 cells [112]. Fabbri et al. identified that miR-21 and miR-29a were significantly up-regulated in exosomes isolated from A549 cells. Exosomal miR-21and miR-29a bound and activated Toll-like receptors (TLRs) in surrounding immune cells, leading to tumor growth and metastasis [113]. Platelet-derived exosomes delivered the glycoprotein IIb/IIIa (CD41) to the surface of lung cancer cells, and induced phosphorylation of MAPK p42/44 and AKT as well as the expression of cyclin D2 in lung cancer cells, facilitating lung cancer cells proliferation [114]. EVs transferred miR-142-3p from lung adenocarcinoma cells to both endothelial and fibroblast cells, which promoted angiogenesis by inhibition of TGFβR1 in endothelial cells. In additional, EV-associated miR-142-3p also facilitated the cancer-associated fibroblast (CAF) transformation but did not target TGFβR1 in lung fibroblast cells [115]. Taken together, these data indicate that exosomes can transfer miRNA from lung cancer cells to other non-cancer cells and regulate gene expression in the host cells through binding to their target mRNAs, moreover, exosomal proteins also directly activate signaling in the host cells, with resultant regulation growth and proliferation of lung cancer.
Exosomes and lung cancer immune escape
Tumor-derived exosomes carry immunosuppressive molecules, and transfer these cargos to immune cells and directly or indirectly suppress the functions of immune cells, thereby promoting tumor progression [116]. Chen et al. reported that exosomes from human melanoma, breast and lung cancer cells carry immunosuppressive PD-L1, and PD-L1 binds to PD-1 via its extracellular domain to inactivate T cells. The level of exosomal PD-L1 was up-regulated by interferon-γ (IFN-γ), which suppressed the function of CD8 T cells and facilitated tumor growth [117]. In addition, Poggio et al. also uncovered the mechanism that exosomal PD-L1 enables tumor cells to extend survival, and genetic blockade of exosomal PD-L1 can facilitate T-cell activity in the draining lymph node, thereby inducing systemic antitumor immunity and memory [118]. Berchem et al. reported that hypoxic tumor-derived microvesicles (TD-MVs) carried immunosuppressive molecules (TGF-β1 and miR-23a) that can inhibit natural killer (NK) cells cytotoxicity in vitro and in vivo [119]. Huang et al. discovered that more than 80% exosomes isolated from the lung cancer biopsies contained EGFR, and dendritic cells (DCs) can capture these exosomes to differentiate into tolerogenic DCs, which could convert the naïve CD4+ T cells into tumor antigen-specific regulatory T cells (Tregs), thus suppressing the function of tumor specific CD8+ T cells [120]. Collectively, these data show that tumor-derived exosomes can rescue tumor cells by evading the surveillance of immune cells, which could represent a therapeutic target, contribute to developing immunotherapeutic approaches for cancer therapy.
Exosomes and lung cancer metastasis
Metastasis is a complex multistep process of tumor cells invasion, survival in blood vessels, attachment to and colonization of the host organs, during which exosomes act as functional mediators in cell–cell communication, influencing the various steps of the metastatic cascades [121]. Hoshino et al. have demonstrated that certain exosomal integrins are ‘addressing’ cancer cells to specific organs and predict organ-specific metastasis, among which exosomal integrins α6β4 and α6β1 were linked to lung metastasis, while exosomal integrin αvβ5 was associated with liver metastasis [122]. Liang et al. demonstrated that platelets from NSCLC patients expressed higher levels of miR-223 than those from healthy subjects. Incubation of A549 cells with platelet-secreted microvesicles suppressed the translation of 3′UTR of erythrocyte membrane protein band 4.1-like 3 (EPB41L3) in A549 cells, promoting A549 cells invasion [123]. Exosomes from pigment epithelium-derived factor (PEDF)-treated A549 cells significantly inhibited cells motility compared to control groups, and it was further found that PEDF decreased cancer-derived exosome-mediated metastatic activity through elevated thrombospondin 1(THBS1) expression and increased its contents in exosomes. Owing to THBS1 suppressed cytoskeletal remodeling and the migration and invasion of lung cancer cells, the presence of THBS1-containing exosomes in the pre-metastatic niche could prevent lung cancer progression [124]. Compared with lung cells, MMP3 proteins were increased in 3T3-L1 adipocytes. 3T3-L1 adipocyte-derived exosomes (3T3-A-EXO) transferred MMP3 to 3LL Lewis lung cancer cells, then, MMP3 induced MMP9 expression in 3LL cells and promoted 3LL cells invasion in vitro and in vivo [125].
In NSCLC, bone metastasis is the frequent complication. Taverna S et al. investigated that exosomes derived from NSCLC cells contain a high level of EGFR binds amphiregulin (AREG), which induce the activation of EGFR signaling in pre-osteoclasts that in turn up-regulates the expression of matrix metalloproteinases 9 (MMP9) and tartrate-resistant acid phosphatase (TRAP), well-known genes involved in osteoclasts differentiation, triggering osteolytic bone metastasis [126]. In addition, Xu et al. demonstrated that miR-21 overexpression in exosomes from A549 cells, and exosomal miR-21 was taken up by osteoclast progenitor cells, inhibited PDCD4 expression in recipient cells, thereby promoting osteoclastogenesis [127].
SCLC is a strong preference for early brain metastases, which lead to most SCLC-related deaths. SCLC cells cocultured with exosomes derived from human brain microvascular endothelial cells (HBMECs) showed that S100A16, a partner of the annexin family of proteins, was significantly up-regulated. Meanwhile, HBMECs exosomes induced translocation of S100A16 in the recipient SCLC cells from the cytoplasm to the nucleus, which triggered the expression of prohibitin (PHB)-1, a protein in the mitochondria inner membrane to maintain mitochondrial membrane potential, facilitating brain metastatic SCLC cells to escape death signals and survive in the brain [128].
Taken together, these findings indicate that tumor-derived exosomes serve as a crosstalk between cancer cells and their local and distant microenvironment, and are capable of mediating organ-specific metastasis, which provide support for Stephen Paget’s “seed-and-soil” hypothesis.
Exosomes and EGFR-TKIs resistance
EGFR-TKIs, containing the first generation of gefitinib and erlotinib, are standard first-line therapy for NSCLC patients who harbor sensitivity EGFR mutations, yet, acquired resistance to EGFR-TKIs is inevitable. Jing et al. reported that gefitinib-resistant H827R cells transferred miR-21 to gefitinib-sensitive HCC827 cells by exosomes, which activated the AKT signaling in gefitinib-sensitive HCC827 cells, thereby leading to HCC827 cells to gefitinib resistance [129]. Zhang et al. identified lncRNA RP11-838N2.4 was increased in erlotinib-resistant cells compared to erlotinib-sensitive HCC827 and HCC4006 cells. Treatment-sensitive cells with exosomes secreted by erlotinib-resistant cells induced erlotinib resistance, whereas the knockdown of lncRNA RP11-838 N2.4 aborted this effect. It was further detected that forkhead box protein O1 (FOXO1) was enriched in the promoter region of lncRNA RP11-838N24, leading to its silencing via the recruitment of histone deacetylase. In addition, the high expression levels of serum exosomal lncRNA RP11-838N2.4 in patients displayed resistance to erlotinib treatment [130]. Choi et al. identified exosomal proteins characteristic of getinitib-resistant PC9R cells and found that these proteins mainly originated from membrane-associated components, cytoskeleton, and plasma membrane. Further analysis suggested the AKT/mTOR signaling were correlated with gefitinib resistance. Moreover, exosome-transferred AKT/mTOR complex to recipient cells was crucial to gefitinib resistance [131]. The expression level of miR-214 was higher in gefitinib-resistant PC9GR cells than gefitinib-sensitive PC9 cells. Exosomes transferred miR-214 from PC9GR cells to PC9 cells, which conferred gefitinib resistance in PC9 cells. PC9GR cell-derived exosomes transfected with miR-214 antagomir reversed gefitinib resistance. Thus, exosomal miR-214 may mediate gefitinib resistance [132]. Lei et al. found that the expression of lncRNA H19 was up-regulated in gefitinib-resistant cells when compared with sensitive parent cells. This further demonstrated that exosomal H19 can induce gefitinib resistance [133]. Altogether, these results reveal an association between EGFR-TKIs resistance and exosomes, which provide a new concept to study EGFR-TKIs resistance, meanwhile, exosomes may be a promising therapeutic target for NSCLC patients with EGFR-TKIs resistance.
Exosomes as diagnostic and prognostic biomarkers in lung cancer
There are no validated techniques for screening of lung cancer other than the low-dose computed tomography (LDCT) [134, 135]. Significantly, liquid biopsy is being developed to serve as a minimally invasive diagnostic and predictive tool for lung cancer [136]. Complement fragment C4d was found to be elevated in biological samples from lung cancer patients, and correlated with poor prognosis, which may be of value for the diagnosis and prognosis of lung cancer at a very early stage [137]. Sozzi et al. demonstrated that plasma microRNA signature classifier (MSC) had predictive, diagnostic, and prognostic value, and MSC may complement LDCT screening via reducing false-positive rates [138]. Montani et al. validated a serum microRNA signature (the miR Test) that might serve as a useful tool for lung cancer screening in a high-risk population [139]. Cohen et al. described a multi-analyte blood test, called cancer SEEK, which could detect both the presence of relatively early cancers and localize the organ of origin of cancers through combined evaluation of the levels of circulating proteins and driver gene mutation [140]. Herein, exosomes represent ideal biomarkers in lung cancer (Table 3), apart from their accessibility and stabilization in most body fluids, which also reflect parental cells signature, as detailed below.
Table 3.
Exosomes as diagnostic and prognostic biomarkers in lung cancer
| Exosomal cargos | Biofluid | Method | Clinical significance | References |
|---|---|---|---|---|
| (1) miRNA | ||||
| miR-96 | Serum | qPCR | High expression was positively associated with metastasis and high-grade lung cancer | [141] |
| miR-30a-3p, miR-181-5p, miR-361-5p, and miR-30e-3p | Plasma | miRNA-seq | Lung adenocarcinoma -specific miRNA | [142] |
| miR-15b-5p, miR-320b, and miR-10b-5p | Plasma | miRNA-seq | Lung squamous cell carcinoma-specific miRNA | [142] |
| miR-21, miR-4257 | Plasma | RT-qPCR | High miR-21 and miR-4257 levels predicted recurrence and worse survival rate | [143] |
| miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203,miR-205, miR-210, miR-212, and miR-214 | Plasma | miRNA microarray | Serve as screening markers for lung adenocarcinoma | [144] |
| miR-126 | Serum | qRT-PCR | High levels associated with early-stage NSCLC patients | [145] |
| miR-200b, miR-200c and miR-141 | Pleural effusions | qRT-PCR | High levels associated with lung adenocarcinoma | [146] |
| miR-222-3p | Plasma | qRT-PCR | A prognostic biomarker for predicting gemcitabine sensitivity in NSCLC patients | [147] |
| miR-375, miR-429 and miR-483-5p | Pleural effusion | qRT-PCR | Distinguished lung adenocarcinoma from tuberculous or other benign lesions | [148] |
| miR-21, miR-205 and miR-155 | Serum | A mathematical model | For early detection of NSCLC | [149] |
| miR-378a, miR-139-5p, miR-379, and miR-200b-5p | Plasma | qRT-PCR | Discriminated between lung adenocarcinomas and carcinomas | [150] |
| miR-30a-3p, miR-151a-5p, miR-200b-5p, miR-100, miR-629 and miR-154-3p | Plasma | qRT-PCR | Distinguished lung adenocarcinoma from granuloma | [128] |
| miR-146a-5p | Serum | qRT-PCR | Predicted the efficacy of cisplatin for NSCLC patients and real-time monitoring drug resistance | [151] |
| miR-30b, miR-30c, miR-103, miR-122, miR-195, miR-203, miR-221, and miR-222 | Plasma | qPCR | All correlated with NSCLC | [152] |
| miR-205-5p, miR-200b | Pleural effusions | RT-PCR | All correlated with lung cancer | [153] |
| miR-10b-5p, miR-21-5p, and miR-23b-3p | Plasma | qPCR | High levels related to poor overall survival | [154] |
| miR-425-3p | Serum | qRT-PCR | High levels correlated with platinum-resistant patients | [155] |
| (2) Proteins | ||||
| LBP | Serum | ELISA | Distinguished between patients with metastatic and non-metastatic NSCLC | [156] |
| CD151, CD171 and TSPAN8 | Plasma | EV array | All correlated with lung cancer; high levels of CD151 correlated with SCLC | [157] |
| CD9, CD81 and CD151 | Plasma | EV Array | All correlated with lung cancer | [158] |
| NY-ESO-1 | Plasma | EV Array | Correlated with overall survival | [159] |
| Tim-3/Galectin-9 | Plasma | ELISA | Positively associated with larger tumor size, advanced stages and more lymph node metastasis as well as distant metastasis | [160] |
| HUWE1, TPM3, SRGN, and THBS1 | Plasma | Mass spectrometry | Differentiated lung adenocarcinoma from controls | [161] |
| LRG1 | Urinary | Western blot | High levels correlated with NSCLC | [162] |
| (3) lncRNA | ||||
| MALAT-1 | Serum | qRT-PCR | Positively correlated with tumor stage and lymphatic metastasis | [163] |
| lncRNA GAS5 | Serum | qRT-PCR | Identified patients with early NSCLC | [164] |
| (4) Lipid | ||||
| Lipids | Plasma | UHR-FTMS | Distinguished early- and late-stage NSCLC patients from healthy individuals | [165] |
PCR Quantitative real-time polymerase chain reaction, LBP lipopolysaccharide-binding proteins, ELISA Enzyme-linked immunosorbent assay, Tim-3 T cell Immunoglobulin- and mucin domain-containing molecule 3, HUWE1 E3 ubiquitin-protein ligase HECT, UBA and WWE domain-containing protein 1, TPM3 actin filament-binding protein tropomyosin alpha-3 chain, SRGN secretory vesicle proteoglycan serglycin, THBS1 adhesive glycoprotein thrombospondin-1, LRG1 the leucine-rich a-2-glycoprotein, UHR-FTMS ultra high-resolution fourier transform mass spectrometry, GAS5 growth arrest-specific transcript 5
Exosomal microRNA
Exosomal miR-96 was expressed at higher levels in serum from lung cancer patients than normal people. Accordingly, a higher exosomal miR-96 level was positively associated with metastasis and high-grade lung cancer [141]. Exosomal miRNAs, adenocarcinoma (AC)-specific miR-30a-3p, miR-181-5p, miR-361-5p and miR-30e-3p, and squamous cell carcinoma (SCC)-specific miR-15b-5p, miR-320b and miR-10b-5p, were isolated from the plasma of lung cancer patients. These exosomal miRNAs can discriminate between AC and SCC as noninvasive biomarkers for early NSCLC diagnosis [142]. The exosomal miR-21 and miR-4257 levels of the NSCLC patients showed a significant increase in those subjects with recurrence compared with those without recurrence and healthy individuals. Further assessing the clinical significance of these miRNAs, exosomal miR-21 showed a significant correlation with tumor size and tumor node metastasis (TNM) stage, and exosomal miR-4257 showed a significant correlation with lymphatic invasion, histological type and TNM stage. In addition, the high expression levels of exosomal miR-21 and miR-4257 were related to worse survival rate [143]. Rabinowits et al. detected 12 specific miRNAs (miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214) in the circulating exosomes, suggesting that circulating exosomal miRNA can act as screening markers for lung adenocarcinoma [144]. Exosomal miR-126 was significantly higher in early-stage NSCLC patients than those at advanced stage, showing exosomal miR-126 as a diagnostics biomarker for NSCLC progression [145]. Hydbring et al. investigated exosomal RNA profiling that could distinguish patients with lung adenocarcinoma from benign inflammatory processes. The results revealed that miR-200 family (miR-200b, miR-200c and miR-141) and the mRNA transcript encoding lipocalin-2 (LCN2) were the most substantially up-regulated in the lung adenocarcinoma group, and further suggested miR-200 family (AUC: 0.95) and LCN2 (AUC: 0.9916) as diagnostic markers in lung cancer liquid biopsies [146]. The high level of circulating exosomal miR-222-3p was associated with poor prognosis following gemcitabine therapy in NSCLC patients [147]. Wang et al. identified 9 exosomal miRNAs (miR-141-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-203a-3p, miR-205-5p, miR-375, miR-429 and miR-483-5p) that were the most abundant in pleural effusion of lung adenocarcinoma when compared to that of tuberculosis or other benign lesions. Furthermore, miR-375, miR-429 and miR-483-5p were validated to be correlated with lung adenocarcinoma [148]. Lai et al. developed a mathematical model to assess exosomal miR-21, miR-205 and miR-155 correlation with tumor volume and showed a combination of exosomal miR-21, miR-205 and miR-155 as diagnostic biomarkers for early screening of NSCLC [149]. Cazzoli et al. analyzed exosomal miRNA from 30 plasma samples, identified 4 microRNAs (miR-378a, miR-139-5p, miR-379 and miR-200b-5p) to discriminate between lung adenocarcinomas and carcinomas. In addition, six microRNAs (miR-30a-3p, miR-151a-5p, miR-200b-5p, miR-100, miR-629 and miR-154-3p) were selected to distinguish lung adenocarcinoma from granuloma [150]. The high level of serum exosomal miR-146a-5p showed the chemosensitivity of NSCLC to cisplatin, and reduced recurrence rates. Thus, exosomal miR-146a-5p can be a prognosis biomarker for NSCLC patients to cisplatin response and real-time detection drug resistance [151]. Giallombardo et al. isolated exosomes from 18 plasma samples, identified a panel of exosomal miRNAs (30b, 30c, 122, 103, 203, 195, 222, 221), all associated with NSCLC [152]. Exosomes were isolated from the pleural effusions that were obtained from patients with lung cancer, pulmonary tuberculosis and pneumonia. Exosomal miR-205-5p and miR-200b were significantly increased in lung cancer patients, while exosomal miR-378i was markedly elevated in pulmonary tuberculosis [153]. The high expression of exosomal miR-10b-5p, miR-21-5p and miR-23b-3p was independently related to poor overall survival (OS) [with hazard ratio (95% confidence interval): 2.22 (1.18–4.16), P = 0.013; 2.12 (1.28–3.49), P = 0.003; 2.42 (1.45–4.04), P = 0.001, respectively] [154]. Yuwen et al. analyzed 170 serum samples from advanced NSCLC patients and found that the expression of exosomal miR-425-3p was increased in platinum-resistant patients, which was associated with poor progression-free survival (PFS) [155].
Exosomal proteins
Lipopolysaccharide-binding proteins (LBP) in serum exosomes were discovered to be significantly distinguished between patients with metastatic and non-metastatic NSCLC. Receiver operating characteristic (ROC) curves showed that exosomal LBP had an area under the curve (AUC) of 0.803 with a specificity of 67% and a sensitivity of 83.1% [156]. Exosomes was purified from the plasma of 431 lung cancer patients and 150 controls and tetraspanins CD151, cell adhesion molecule CD171 and TSPAN8 were detected to be significantly up-regulated in the lung cancer patients. However, only CD151 was remarkably up-regulated in the SCLC patients [157]. Jakobsen et al. explored the potential of plasma exosomal protein markers in diagnosing NSCLC. The EV array shows that CD9, CD81 and CD151 were highly expressed in the cancerous patients [158]. Likewise, according to the EV array, NY-ESO-1 showed a strong impact on OS [hazard rate (HR) 1.78, 95% (1.78–2.44); P = 0.0001] in NSCLC [159]. Gao et al. revealed that exosomal Tim-3/Galectin-9 was positively associated with a larger tumor size, advanced stages and more lymph node metastasis as well as distant metastasis. Additionally, exosomes from lung adenocarcinoma showed a lower Tim-3/Galectin-9 compared to those from lung squamous cell carcinoma [160]. Vykoukal et al. analyzed plasma exosomal proteins from subjects with lung adenocarcinoma and matched controls, and identified four exosome-associated proteins, E3 ubiquitin protein ligase HECT, UBA and WWE domain-containing protein 1 (HUWE1), actin filament-binding protein tropomyosin alpha-3 chain (TPM3), secretory vesicle proteoglycan serglycin (SRGN) and adhesive glycoprotein thrombospondin-1 (THBS1) that differentiated lung adenocarcinoma from controls (AUC: 0.90) [161]. The leucine-rich a-2-glycoprotein (LRG1) was validated to be expressed at higher levels in urinary exosomes from NSCLC patients [162].
Exosomal lncRNA
Zhang et al. isolated exosomes from serum of NSCLC patients and healthy subjects and discovered a lncRNA, named MALAT-1, MALAT-1 was significantly increased in exosomes from NSCLC patients, moreover, the level of exosomal MALAT-1 was positively correlated with tumor stage and lymphatic metastasis [163]. In addition, in vitro studies indicated that serum exosome-derived MALAT-1 facilitated tumor growth and metastasis, and reduced lung cancer cells apoptosis [131]. Li et al. found that long noncoding RNA growth arrest-specific transcript 5 (GAS5) was down-regulated in exosomes from NSCLC patients compared with healthy controls. Furthermore, patients with NSCLC in early stage had a higher Exo-GAS5 expression than advanced-stage NSCLC, thus, Exo-GAS5 may be as a noninvasive serum-based marker to identify patients with early NSCLC [164].
Exosomal lipid
Fan et al. explored the lipid profiles of plasma exosomes from 39 normal subjects and 91 NSCLC patients (44 early stage and 47 late stage), using least absolute shrinkage and selection operator (LASSO) and random forests (RF) identified exosomal lipids features that successfully distinguished early- and late-stage NSCLC patients from healthy individuals. The area under the receiver operating characteristic curve for early- and late-stage NSCLC versus healthy individuals using the selected lipid features was 0.79 and 0.77 for LASSO and 0.85 and 0.88 for RF, respectively [165].
The role of exosomes in lung cancer therapy
Exosome-based immunotherapy
There are increasing evidences showing that tumor cell-derived exosomes (TEXs) and dendritic cell-derived exosomes (DEXs) can induce specific antitumor immunity. DEXs have the potential to eradicate established tumors in a T-cell-dependent manner and TEX bear neoantigens internalized by DCs which could trigger antitumor T cell response [166, 167]. A549 cells transfected a Rab27a overexpression vector and exosomes were isolated from those displayed high levels of CD9, CD63, Hsp70 and Hsp90. Subsequently, dendritic cells (DCs) incubated with these exosomes expressed higher levels of MHC class II, CD80 and CD86, which significantly enhanced CD4+ T-cell proliferation in vitro. In addition, it was also demonstrated that these exosomes which were injected into BALB/c mice significantly inhibited tumor growth in vivo [168]. Exosomes from CD40 ligand gene-modified 3LL Lewis lung cancer cells were demonstrated to be more immunogenic. Coculturing CD40 exosomes with DCs revealed that CD40L exosomes induced the maturation of DCs and stimulated tumor antigen-specific CD4+ T cell proliferation, triggering a strong protective antitumor immunity in vivo [169].
Most notably, DEX-based phase I and II clinical trials have been performed in NSCLC, indicating the safety and feasibility of the approach, as well as the preference of these nanovesicles to stimulate T cell- and NK cell-based immune responses in patients [167]. Besse et al. performed a phase II clinical trial assessing the clinical benefit of exosomes derived from TLR4L-or interferon (IFN)-γ-maturated DCs (IFN-γ-DEX) loaded with MHC I- and II-restricted cancer antigens as maintenance immunotherapy after first-line chemotherapy in NSCLC. The median PFS was 2.2 months and median OS was 15 months. 32% of patients had stable disease beyond 4 months. Only one patient displayed a grade 3 hepatotoxicity [30]. Morse et al. performed a phase I study to evaluate the safety, feasibility and efficacy of autologous DC-derived exosomes (DEX) loaded with the MAGE tumor antigens in patients with advanced NSCLC. DEX therapy was well tolerated and some patients had a relatively long PFS and MAGE-specific T cell responses was detected in 3/9 patients [29]. DEX-based phase I and II clinical trials were discovered to be well tolerated in lung cancer patients, yet, because of the small sample, the data lack credibility, so more extensive clinical trials need to be conducted.
Exosomes as anticancer drug delivery vehicles
There are emerging evidences showing that exosomes is a promising drug delivery system, because exosomes have unique characteristics, for instance, low immunogenicity, decreased toxic, high biocompatibility, and surmount biological barriers (including blood–brain barrier), nano-scale size and target homing specificity [170, 171]. Sun et al. reported a novel nanoparticle drug delivery system, exosome–curcumin complexes, and showed that exosomes can increase solubility, stability, and bioavailability of curcumin, thus enhancing the anti-inflammatory activity of curcumin [172]. Maguire et al. demonstrated that microvesicle-associated adeno-associated virus (AAV) vector as a novel gene delivery system could enhance gene transfer in culture cells [173]. Mizrak et al. evaluated that MVs can act as a cell-derived gene delivery vehicle transferring therapeutic mRNA/protein to target tumor cells, which inhibited schwannoma tumor growth through expressing a high level of functional protein in recipient cells [174]. Tian et al. validated a doxorubicin delivery platform using exosomes that showed highly antitumor activity both in vitro and in vivo [175]. In addition, Kim et al. assessed the feasibility of an exosome-based paclitaxel (PTX) formulation treating multiple drug resistance (MDR) cancer. The result showed that the incorporation of PTX into exosomes significantly increased drug accumulation levels and cytotoxicity in resistant cells. Furthermore, Kim et al. demonstrated that airway administered exosomes arrived to pulmonary metastases and transported their drug payload to target cancer cells in vivo [176]. Srivastava et al. developed an exosome-based drug delivery system for lung cancer therapy, called nanosomes, which consists of GNPs conjugated to anticancer drug doxorubicin (Dox) via a pH-sensitive hydrazone linker loaded onto the exosomes (Exo-GNP-Dox). The increased rate of drug release under acidic conditions, successful uptake of the nanosomes by the recipient cells and the cell viability analysis indicated that nanosomes displayed preferential cytotoxicity to cancer cells other than non-cancerous cells [31]. Celastrol, a plant-derived triterpenoid, suppressed the proliferation of A549 and H1299 cells by its repressive effects on NF-κB activation and cell cycle progression and also through the induction of chronic endoplasmic reticulum stress-mediated apoptosis. Encapsulation of celastrol into bovine milk-derived exosomes showed greater antitumor efficacy and reduced toxic side effects compared to free celastrol [32]. Kim et al. developed an exosome-based drug delivery system AA-PEG-exoPTX that constitutes paclitaxel (PTX)-loaded macrophage-derived exosomes (exo) with incorporated aminoethylanisamide-polyethylene glycol (AA-PEG) vector moiety. It was further demonstrated that AA-PEG-exoPTX AA-PEG-exoPTX efficiently inhibited lung metastases through targeting the sigma receptor that was overexpressed in lung cancer cells [33]. These data supported that exosome-based drug formulations can provide powerful and low immunogenic profile delivery systems for anticancer therapy, however, there are some technological, functional and safe issues that remain unresolved, such as preferential accumulation in cancer cells, efficient transportation of incorporated cargo into target cancer cells and controlled drug release, so more researches are necessary to unveil these mechanisms.
Conclusion
In summary, exosomes can influence lung cancer progression, and exosomes may be an alternative option for lung cancer therapy. Nevertheless, some problems are yet to be addressed. First, bulk extraction and thorough characterization of exosomes remain challenging, which will influence clinical applications, biomedical investigation as well as the production cost, thus, we could combine multiple separation technologies to optimize exosomes isolation and enrichment. Second, exosomes can transfer biologically active molecules between lung cancer cells and their microenvironment (Fig. 2 and Table 2), but, the mechanisms exosomes use to exactly deliver certain cargos to specific target cells need to be illuminated. Third, exosomes are a promising biomarker for lung cancer diagnosis, prediction and prognosis as well as real-time monitoring therapy response (Table 3). However, these studies are generally found with small samples and poor repeatability, so large multicenter studies are necessary to develop effectiveness of liquid biopsy. Finally, the exosome-based clinical trials have been accomplished in lung cancer, moreover, there are still some clinical trials going on (Table 4) and such studies can provide proof for the transition of therapeutics that are based on exosomes into the clinic in the not-so-distant future (Fig. 3).
Table 4.
Current clinical trials on exosomes in lung cancer
| Title | Status | Conditions | Interventions |
|---|---|---|---|
| Combined diagnosis of CT and exosome in early lung cancer | Not yet recruiting | Early lung cancer | Procedure: surgery |
| Serum exosomal long noncoding RNAs potential biomarkers for lung cancer diagnosis | Recruiting | Lung cancer | Diagnostic test: collect samples |
| Clinical research for the consistency analysis of PD-L1 in lung cancer tissue and plasma exosome before and after radiotherapy | Unknown | NSCLC | Radiation: radiotherapy |
| Dynamic monitoring circulating tumor DNA in surgical patients with lung cancer | Recruiting | Lung cancer | Other: blood samples |
| Detection of circulating biomarkers of immunogenic cell death | Active, not recruiting | NSCLC | Other: blood samples |
| Circulating exosome RNA in lung metastases of primary high grade | Recruiting | Lung metastases | |
| Osteosarcoma | Other: blood samples | ||
| Olmutinib trial in T790 M (+) NSCLC patients detected by liquid biopsy using BALF extracellular vesicular DNA | Active, not recruiting | NSCLC | Drug: olmutinib |
| Detection of either the EML4-ALK gene rearrangements or the T790 M EGFR mutation in the plasma of advanced NSCLC patients | Active, not recruiting | Carcinoma | |
| NSCLC | Other: blood samples | ||
| Pilot study with the aim to quantify a stress protein in the blood and in the urine for the monitoring and early diagnosis of malignant solid tumor | Recruiting | Cancer | Other: blood samples |
| Other: urine samples |
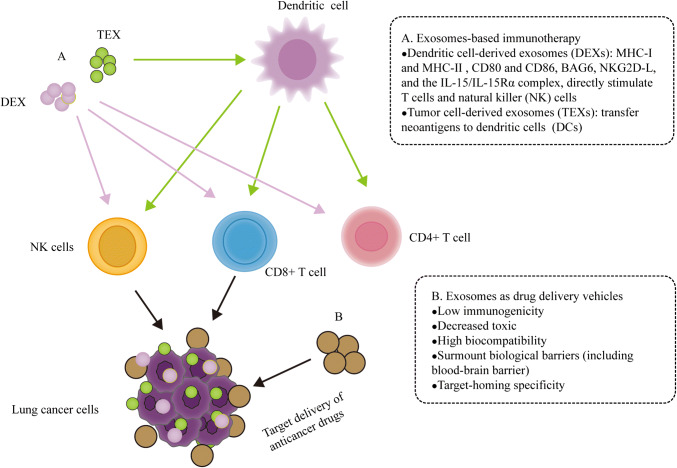
Fig. 3.
The role of exosomes in lung cancer therapy. a. Tumor cell-derived exosomes (TEXs) and dendritic cell-derived exosomes (DEXs) can induce specific antitumor immunity. TEXs carry neoantigens and deliver immunosuppressive cargos to dendritic cells (DCs), which influence the development, maturation and antitumor activities of immune cells. The presence of MHC I, MHC II molecules and costimulatory molecules (CD80 and CD86) as well as other molecules on the surface of DEX makes them directly stimulate T cells and natural killer (NK) cells, triggering a strong immunity response [166, 167]. b. Exosome-based drug vehicles have unique characteristics, such as low immunogenicity, decreased toxic, high biocompatibility, surmount biological barriers and target homing specificity, which make them provide powerful and low immunogenic profile delivery systems for anticancer therapy [170, 171]
Acknowledgements
The study was supported by Natural Science Foundation of China (Grants 81472124, 81774291 to Yongchun Yu, and 81573890 to Jianli Sun).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianli Sun, Email: 1721679167@qq.com.
Xiao Zhang, Email: 386229418@qq.com.
Yongchun Yu, Email: yuyongchun1255@126.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Herbst Roy S, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd FA, Pereira JR, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, Mv Kooten, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabárbara P, Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang J-J, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Park K, Tan E-H, O’Byrne K, Zhang L, Boyer M, Mok T, Hirsh V, Yang JC-H, Lee KH, Lu S, Shi Y, Kim S-W, Laskin J, Kim D-W, Arvis CD, Kölbeck K, Laurie SA, Tsai C-M, Shahidi M, Kim M, Massey D, Zazulina V, Paz-Ares L. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17(5):577–589. doi: 10.1016/s1470-2045(16)30033-x. [DOI] [PubMed] [Google Scholar]
- 7.Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, Okamoto I, Zhou C, Cho BC, Cheng Y, Cho EK, Voon PJ, Planchard D, Su W-C, Gray JE, Lee S-M, Hodge R, Marotti M, Rukazenkov Y, Ramalingam SS. Osimertinib in untreated EGFR-mutated advanced non-small-cell Lung cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 8.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 10.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, Lee JS, Hellmann MD, Hamid O, Goldman JW, Soria JC, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L, Investigators K- Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 11.Herbst RS, Baas P, Kim D-W, Felip E, Pérez-Gracia JL, Han J-Y, Molina J, Kim J-H, Arvis CD, Ahn M-J, Majem M, Fidler MJ, de Castro G, Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M, Garon EB. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540–1550. doi: 10.1016/s0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 12.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR, Investigators K- Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 13.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Aren Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhaufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crino L, Blumenschein GR, Jr, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi NA, Mazières J, Planchard D, Stinchcombe TE, Dy GK, Antonia SJ, Horn L, Lena H, Minenza E, Mennecier B, Otterson GA, Campos LT, Gandara DR, Levy BP, Nair SG, Zalcman G, Wolf J, Souquet P-J, Baldini E, Cappuzzo F, Chouaid C, Dowlati A, Sanborn R, Lopez-Chavez A, Grohe C, Huber RM, Harbison CT, Baudelet C, Lestini BJ, Ramalingam SS. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16(3):257–265. doi: 10.1016/s1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, Braiteh F, Waterkamp D, He P, Zou W, Chen DS, Yi J, Sandler A, Rittmeyer A. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387(10030):1837–1846. doi: 10.1016/s0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 17.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, Cortinovis DL, Leach J, Polikoff J, Barrios C, Kabbinavar F, Frontera OA, De Marinis F, Turna H, Lee J-S, Ballinger M, Kowanetz M, He P, Chen DS, Sandler A, Gandara DR. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255–265. doi: 10.1016/s0140-6736(16)32517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 19.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bullock MD, Silva AM, Kanlikilicer-Unaldi P, Filant J, Rashed MH, Sood AK, Lopez-Berestein G, Calin GA. Exosomal non-coding RNAs: diagnostic, prognostic and therapeutic applications in cancer. Noncoding RNA. 2015;1(1):53–68. doi: 10.3390/ncrna1010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnstone RM, Adam M, Hammon JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 22.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ. Reassessment of exosome composition. Cell. 2019;177(2):428–445e418. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91(4):431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ung TH, Madsen HJ, Hellwinkel JE, Lencioni AM, Graner MW. Exosome proteomics reveals transcriptional regulator proteins with potential to mediate downstream pathways. Cancer Sci. 2014;105(11):1384–1392. doi: 10.1111/cas.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 27.Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Hoen ENN-t, Piper MG, Sivaraman S, Skog J, Théry C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2(1):20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014 doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Valente N, Shreeniwas R, Sutton MA, Delcayre A, Hsu DH, Le Pecq JB, Lyerly HK. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3(1):9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, Le Chevalier T, Livartoski A, Barlesi F, Laplanche A, Ploix S, Vimond N, Peguillet I, Thery C, Lacroix L, Zoernig I, Dhodapkar K, Dhodapkar M, Viaud S, Soria JC, Reiners KS, Pogge von Strandmann E, Vely F, Rusakiewicz S, Eggermont A, Pitt JM, Zitvogel L, Chaput N. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 2016;5(4):e1071008. doi: 10.1080/2162402X.2015.1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava A, Amreddy N, Babu A, Panneerselvam J, Mehta M, Muralidharan R, Chen A, Zhao YD, Razaq M, Riedinger N, Kim H, Liu S, Wu S, Abdel-Mageed AB, Munshi A, Ramesh R. Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci Rep. 2016;6:38541. doi: 10.1038/srep38541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aqil F, Kausar H, Agrawal AK, Jeyabalan J, Kyakulaga AH, Munagala R, Gupta R. Exosomal formulation enhances therapeutic response of celastrol against lung cancer. Exp Mol Pathol. 2016;101(1):12–21. doi: 10.1016/j.yexmp.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL, Kabanov AV, Batrakova EV. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine. 2018;14(1):195–204. doi: 10.1016/j.nano.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Ela S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 35.Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol. 2017;13(12):731–749. doi: 10.1038/nrneph.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, Lotvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Colao IL, Corteling R, Bracewell D, Wall I. Manufacturing exosomes: a promising therapeutic platform. Trends Mol Med. 2018;24(3):242–256. doi: 10.1016/j.molmed.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 41.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 42.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16(4):415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 44.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 46.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458(7235):172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 50.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 51.Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9(1):40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 53.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 54.Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, Kuchimanchi S, Peng CG, Ruda VM, Del Conte-Zerial P, Hengstler JG, Kalaidzidis Y, Koteliansky V, Zerial M. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485(7399):465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
- 55.Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284(18):12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu C, Morohashi Y, Yoshimura S-i, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Gronborg M, Bius WM, Rhee J, Barr FA, Simons M. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2014;189(2):223–232. doi: 10.1083/icb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 58.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Squadrito ML, Baer C, Fdr Burdet, Maderna C, Gilfillan GD, Lyle R, Ibberson M, Palma MD. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 61.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 62.Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 63.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450(7168):435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 65.Denzer K, Mv Eijk, Kleijmeer MJ, Jakobson E, Groot Cd, Geuze HJ. Follicular dendritic cells carry MHC Class II-expressing microvesicles at their surface. J Immunol Methods. 2000;165(3):1259–1265. doi: 10.4049/jimmunol.165.3.1259. [DOI] [PubMed] [Google Scholar]
- 66.Damke H, Baba T, Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 2014;131(1):69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem. 2014;289(32):22258–22267. doi: 10.1074/jbc.M114.588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci Rep. 2015;5:10300. doi: 10.1038/srep10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11(5):675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 70.Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem. 2010;111(2):488–496. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 71.Théry C, Clayton A, Amigorena S, Ga Raposo. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;3(22):1–29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 72.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118(4):1917–1950. doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2015;199:145–155. doi: 10.1016/j.jconrel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen L-AA, Meerbeeck JP, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31(1):114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 75.Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles. 2014 doi: 10.3402/jev.v3.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82(9):1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 77.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Claytona A, Courta J, Navabia H, Adamsa M, Mason MD, Hobot JA, Newman GR, Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247(1–2):163–174. doi: 10.1016/S0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 79.Lamparski HG, Metha-Damani A, Yao J-Y, Patel S, Hsu D-H, Ruegg C, Pecq J-BL. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 80.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Moller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. 2014 doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhaes A, Ferreira JA, Osorio H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, Lyden D. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reategui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, Floyd FP, Jr, Khankhel AH, Thapar V, Hochberg FH, Sequist LV, Nahed BV, Carter BS, Toner M, Balaj L, Ting TD, Breakefield XO, Stott SL. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat Commun. 2018;9(1):175. doi: 10.1038/s41467-017-02261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shelke GV, Lasser C, Gho YS, Lotvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014 doi: 10.3402/jev.v3.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 86.Yuana Y, Koning RI, Kuil ME, Rensen PC, Koster AJ, Bertina RM, Osanto S. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J Extracell Vesicles. 2013;2:1. doi: 10.3402/jev.v2i0.21494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma S, Rasoo HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4(4):1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ, Harrison P, Sargent IL. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7(6):780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu Y, Nakane N, Maurer-Spurej E. Novel test for microparticles in platelet-rich plasma and platelet concentrates using dynamic light scattering. Transfusion. 2011;51(2):363–370. doi: 10.1111/j.1537-2995.2010.02819.x. [DOI] [PubMed] [Google Scholar]
- 90.Coumans FA, van der Pol E, Boing AN, Hajji N, Sturk G, van Leeuwen TG, Nieuwland R. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J Extracell Vesicles. 2014;3:25922. doi: 10.3402/jev.v3.25922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khodashenas S, Khalili S, Forouzandeh Moghadam M. A cell ELISA based method for exosome detection in diagnostic and therapeutic applications. Biotechnol Lett. 2019;41(4–5):523–531. doi: 10.1007/s10529-019-02667-5. [DOI] [PubMed] [Google Scholar]
- 92.Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, Ivanov AR. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J Proteome Res. 2015;14(6):2367–2384. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 93.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, Carter B, Breakefield XO, Krichevsky AM. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8(1):1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 100.Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, Oeh J, Modrusan Z, Bais C, Sampath D, Ferrara N. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Al-Nedawi K, Meehana B, Kerbelb RS, Allisonc AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. PNAS. 2009;106(10):3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, Wu CY, Kuo PL. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36(34):4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 103.Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X, Shi L, Lu X, Xu W, Lu L, Qin Y, Xiang Q, Liu Q. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–135. doi: 10.1016/j.canlet.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 104.Wu DM, Deng SH, Liu T, Han R, Zhang T, Xu Y. TGF-beta-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 2018;7(10):5118–5129. doi: 10.1002/cam4.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cui H, Seubert B, Stahl E, Dietz H, Reuning U, Moreno-Leon L, Ilie M, Hofman P, Nagase H, Mari B, Kruger A. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene. 2015;34(28):3640–3650. doi: 10.1038/onc.2014.300. [DOI] [PubMed] [Google Scholar]
- 106.Greening DW, Gopala SK, Mathiasa RA, Liub L, Shengb J, Zhub H-J, Simpsona RJ. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin Cell Dev Biol. 2015;40:60–71. doi: 10.1016/j.semcdb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 107.Blackwell RH, Foreman KE, Gupta GN. The role of cancer-derived exosomes in tumorigenicity and epithelial-to-mesenchymal Transition. Cancers (Basel) 2017 doi: 10.3390/cancers9080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol Sci. 2016;37(7):606–617. doi: 10.1016/j.tips.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 109.Kim J, Kim TY, Lee MS, Mun JY, Ihm C, Kim SA. Exosome cargo reflects TGF-beta1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem Biophys Res Commun. 2016;478(2):643–648. doi: 10.1016/j.bbrc.2016.07.124. [DOI] [PubMed] [Google Scholar]
- 110.Rahman MA, Barger JF, Lovat F, Gao M, Otterson GA, Nana-Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016;7(34):54852–54866. doi: 10.18632/oncotarget.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang S, Li X, Zhu R, Han Q, Zhao RC. Lung cancer exosomes initiate global long non-coding RNA changes in mesenchymal stem cells. Int J Oncol. 2016;48(2):681–689. doi: 10.3892/ijo.2015.3272. [DOI] [PubMed] [Google Scholar]
- 112.Xiao H, Lasser C, Shelke GV, Wang J, Radinger M, Lunavat TR, Malmhall C, Lin LH, Li J, Li L, Lotvall J. Mast cell exosomes promote lung adenocarcinoma cell proliferation role of KIT-stem cell factor signaling. Cell Commun Signal. 2014;12:64. doi: 10.1186/s12964-014-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fabbria M, Paonea A, Calorea F, Gallia R, Gaudioa E, Santhanama R, Lovata F, Faddaa P, Maoa C, Nuovob GJ, Zanesia N, Crawfordc M, Ozera GH, Wernickea D, Aldera H, Caligiurid MA, Nana-Sinkamc P, Perrottia D, Crocea CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. PNAS. 2012;109(31):2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, Ratajczak MZ. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113(5):752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 115.Lawson J, Dickman C, Towle R, Jabalee J, Javer A, Garnis C. Extracellular vesicle secretion of miR-142-3p from lung adenocarcinoma cells induces tumor promoting changes in the stroma through cell-cell communication. Mol Carcinog. 2018 doi: 10.1002/mc.22935. [DOI] [PubMed] [Google Scholar]
- 116.Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126(4):1216–1223. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177(2):414–427e413. doi: 10.1016/j.cell.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le Cam E, Nanbakhsh A, Moussay E, Mami-Chouaib F, Janji B, Chouaib S. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-beta and miR23a transfer. Oncoimmunology. 2016;5(4):e1062968. doi: 10.1080/2162402X.2015.1062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang SH, Li Y, Zhang J, Rong J, Ye S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 2013;31(5):330–335. doi: 10.3109/07357907.2013.789905. [DOI] [PubMed] [Google Scholar]
- 121.Steinbichler TB, Dudas J, Riechelmann H, Skvortsova II. The role of exosomes in cancer metastasis. Semin Cancer Biol. 2017;44:170–181. doi: 10.1016/j.semcancer.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 122.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L, Liu Y, Chen X, Zhang CY, Gu H, Zen K. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer. 2015;14:58. doi: 10.1186/s12943-015-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang WT, Chong IW, Chen HL, Li CY, Hsieh CC, Kuo HF, Chang CY, Chen YH, Liu YP, Lu CY, Liu YR, Liu PL. Pigment epithelium-derived factor inhibits lung cancer migration and invasion by upregulating exosomal thrombospondin 1. Cancer Lett. 2018;442:287–298. doi: 10.1016/j.canlet.2018.10.031. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Wu Y, Guo J, Fei X, Yu L, Ma S. Adipocyte-derived exosomes promote lung cancer metastasis by increasing MMP9 activity via transferring MMP3 to lung cancer cells. Oncotarget. 2017;8(47):81880–81891. doi: 10.18632/oncotarget.18737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Taverna S, Pucci M, Giallombardo M, Di Bella MA, Santarpia M, Reclusa P, Gil-Bazo I, Rolfo C, Alessandro R. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci Rep. 2017;7(1):3170. doi: 10.1038/s41598-017-03460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu Z, Liu X, Wang H, Li J, Dai L, Li J, Dong C. Lung adenocarcinoma cell-derived exosomal miR-21 facilitates osteoclastogenesis. Gene. 2018;666:116–122. doi: 10.1016/j.gene.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 128.Xu ZH, Miao ZW, Jiang QZ, Gan DX, Wei XG, Xue XZ, Li JQ, Zheng F, Qin XX, Fang WG, Chen YH, Li B. Brain microvascular endothelial cell exosome-mediated S100A16 upregulation confers small-cell lung cancer cell survival in brain. FASEB J. 2018;33(2):1742–1757. doi: 10.1096/fj.201800428r. [DOI] [PubMed] [Google Scholar]
- 129.Jing C, Cao H, Qin X, Yu S, Wu J, Wang Z, Ma R, Feng J. Exosome-mediated gefitinib resistance in lung cancer HCC827 cells via delivery of miR-21. Oncol Lett. 2018;15(6):9811–9817. doi: 10.3892/ol.2018.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang W, Cai X, Yu J, Lu X, Qian Q, Qian W. Exosome-mediated transfer of lncRNA RP11838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53(2):527–538. doi: 10.3892/ijo.2018.4412. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131.Choi DY, You S, Jung JH, Lee JC, Rho JK, Lee KY, Freeman MR, Kim KP, Kim J. Extracellular vesicles shed from gefitinib-resistant nonsmall cell lung cancer regulate the tumor microenvironment. Proteomics. 2014;14(16):1845–1856. doi: 10.1002/pmic.201400008. [DOI] [PubMed] [Google Scholar]
- 132.Zhang Y, Li M, Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem Biophys Res Commun. 2018;507(1–4):457–464. doi: 10.1016/j.bbrc.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 133.Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol Rep. 2018;40(6):3438–3446. doi: 10.3892/or.2018.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Team NLSTR, Church TR, Black WC, Aberle DR, Berg CD, Clingan KL, Duan F, Fagerstrom RM, Gareen IF, Gierada DS, Jones GC, Mahon I, Marcus PM, Sicks JD, Jain A, Baum S. Results of initial low-dose computed tomographic screening for lung cancer. N Engl J Med. 2013;368(21):1980–1991. doi: 10.1056/NEJMoa1209120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Team TNLSTR. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Levy B, Hu ZI, Cordova KN, Close S, Lee K, Becker D. Clinical utility of liquid diagnostic platforms in non-small cell lung cancer. Oncologist. 2016;21(9):1121–1130. doi: 10.1634/theoncologist.2016-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ajona D, Pajares MJ, Corrales L, Perez-Gracia JL, Agorreta J, Lozano MD, Torre W, Massion PP, de-Torres JP, Jantus-Lewintre E, Camps C, Zulueta JJ, Montuenga LM, Pio R. Investigation of complement activation product c4d as a diagnostic and prognostic biomarker for lung cancer. J Natl Cancer Inst. 2013;105(18):1385–1393. doi: 10.1093/jnci/djt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, Roz L, Conte D, Grassi M, Sverzellati N, Marchiano A, Negri E, La Vecchia C, Pastorino U. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32(8):768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, Bertolotti R, Bellomi M, Rampinelli C, Maisonneuve P, Spaggiari L, Veronesi G, Nicassio F, Di Fiore PP, Bianchi F. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst. 2015;107(6):djv063. doi: 10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- 140.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA, Jr, Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B, Xie Y, Ye Y, Liu J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med. 2017;21(6):1228–1236. doi: 10.1111/jcmm.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, Liu L, Lin B, Su H, Zhao L, Su M, Pan H, Shen L, Xie D, Xie C. Evaluation of Tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 143.Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett. 2017;13(3):1256–1263. doi: 10.3892/ol.2017.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/clc.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 145.Grimolizzi F, Monaco F, Leoni F, Bracci M, Staffolani S, Bersaglieri C, Gaetani S, Valentino M, Amati M, Rubini C, Saccucci F, Neuzil J, Tomasetti M, Santarelli L. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep. 2017;7(1):15277. doi: 10.1038/s41598-017-15475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hydbring P, De Petris L, Zhang Y, Branden E, Koyi H, Novak M, Kanter L, Haag P, Hurley J, Tadigotla V, Zhu B, Skog J, Viktorsson K, Ekman S, Lewensohn R. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer. 2018;124:45–52. doi: 10.1016/j.lungcan.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 147.Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N, Li Y, Liu Y. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16(1):132. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Y, Xu YM, Zou YQ, Lin J, Huang B, Liu J, Li J, Zhang J, Yang WM, Min QH, Li SQ, Gao QF, Sun F, Chen QG, Zhang L, Jiang YH, Deng LB, Wang XZ. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine (Baltimore) 2017;96(44):e8361. doi: 10.1097/MD.0000000000008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lai X, Friedman A. Exosomal miRs in lung cancer: a mathematical model. PLoS ONE. 2016;11(12):e0167706. doi: 10.1371/journal.pone.0167706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. miR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2650–2658. [PubMed] [Google Scholar]
- 152.Giallombardo M, Chacartegui Borras J, Castiglia M, Van Der Steen N, Mertens I, Pauwels P, Peeters M, Rolfo C. Exosomal miRNA analysis in non-small cell lung cancer (NSCLC) patients’ plasma through qPCR: a feasible liquid biopsy tool. J Vis Exp. 2016 doi: 10.3791/53900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin J, Wang Y, Zou YQ, Chen X, Huang B, Liu J, Xu YM, Li J, Zhang J, Yang WM, Min QH, Sun F, Li SQ, Gao QF, Wang XZ. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumour Biol. 2016 doi: 10.1007/s13277-016-5410-6. [DOI] [PubMed] [Google Scholar]
- 154.Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, Xiang Y, Wu N, Wu L, Bai L, Li Y. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8(8):13048–13058. doi: 10.18632/oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yuwen D, Ma Y, Wang D, Gao J, Li X, Xue W, Fan M, Xu Q, Shen Y, Shu Y. Prognostic role of circulating exosomal miR-425-3p for the response of NSCLC to platinum-based chemotherapy. Cancer Epidemiol Biomarkers Prev. 2018 doi: 10.1158/1055-9965.epi-18-0569. [DOI] [PubMed] [Google Scholar]
- 156.Wang N, Song X, Liu L, Niu L, Wang X, Song X, Xie L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018;109(5):1701–1709. doi: 10.1111/cas.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sandfeld-Paulsen B, Jakobsen KR, Baek R, Folkersen BH, Rasmussen TR, Meldgaard P, Varming K, Jorgensen MM, Sorensen BS. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. 2016;11(10):1701–1710. doi: 10.1016/j.jtho.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 158.Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, Jorgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659. doi: 10.3402/jev.v4.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sandfeld-Paulsen B, Aggerholm-Pedersen N, Baek R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR, Varming K, Jorgensen MM, Sorensen BS. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol. 2016;10(10):1595–1602. doi: 10.1016/j.molonc.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gao J, Qiu X, Li X, Fan H, Zhang F, Lv T, Song Y. Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochem Biophys Res Commun. 2018;498(3):409–415. doi: 10.1016/j.bbrc.2018.02.114. [DOI] [PubMed] [Google Scholar]
- 161.Vykoukal J, Sun N, Aguilar-Bonavides C, Katayama H, Tanaka I, Fahrmann JF, Capello M, Fujimoto J, Aguilar M, Wistuba II, Taguchi A, Ostrin EJ, Hanash SM. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget. 2017;8(56):95466–95480. doi: 10.18632/oncotarget.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32(15):1976–1983. doi: 10.1002/elps.201000598. [DOI] [PubMed] [Google Scholar]
- 163.Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490(2):406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 164.Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y, Fan H, Lv T, Liu H, Song Y. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019 doi: 10.1002/jcp.28678. [DOI] [PubMed] [Google Scholar]
- 165.Fan TWM, Zhang X, Wang C, Yang Y, Kang WY, Arnold S, Higashi RM, Liu J, Lane AN. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal Chim Acta. 2018;1037:256–264. doi: 10.1016/j.aca.2018.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Syn NL, Wang L, Chow EK, Lim CT, Goh BC. Exosomes in cancer nanomedicine and immunotherapy: prospects and challenges. Trends Biotechnol. 2017;35(7):665–676. doi: 10.1016/j.tibtech.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 167.Pitt JM, Andre F, Amigorena S, Soria JC, Eggermont A, Kroemer G, Zitvogel L. Dendritic cell-derived exosomes for cancer therapy. J Clin Invest. 2016;126(4):1224–1232. doi: 10.1172/JCI81137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li W, Mu D, Tian F, Hu Y, Jiang T, Han Y, Chen J, Han G, Li X. Exosomes derived from Rab27a overexpressing tumor cells elicit efficient induction of antitumor immunity. Mol Med Rep. 2013;8(6):1876–1882. doi: 10.3892/mmr.2013.1738. [DOI] [PubMed] [Google Scholar]
- 169.Wang J, Wang L, Lin Z, Tao L, Chen M. More efficient induction of antitumor T cell immunity by exosomes from CD40L gene-modified lung tumor cells. Mol Med Rep. 2014;9(1):125–131. doi: 10.3892/mmr.2013.1759. [DOI] [PubMed] [Google Scholar]
- 170.Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 172.Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D, Zhang HG. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18(9):1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maguire CA, Balaj L, Sivaraman S, Crommentuijn MH, Ericsson M, Mincheva-Nilsson L, Baranov V, Gianni D, Tannous BA, Sena-Esteves M, Breakefield XO, Skog J. Microvesicle-associated AAV vector as a novel gene delivery system. Mol Ther. 2012;20(5):960–971. doi: 10.1038/mt.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mizrak A, Bolukbasi MF, Ozdener GB, Brenner GJ, Madlener S, Erkan EP, Strobel T, Breakefield XO, Saydam O. Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Mol Ther. 2013;21(1):101–108. doi: 10.1038/mt.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 176.Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12(3):655–664. doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]