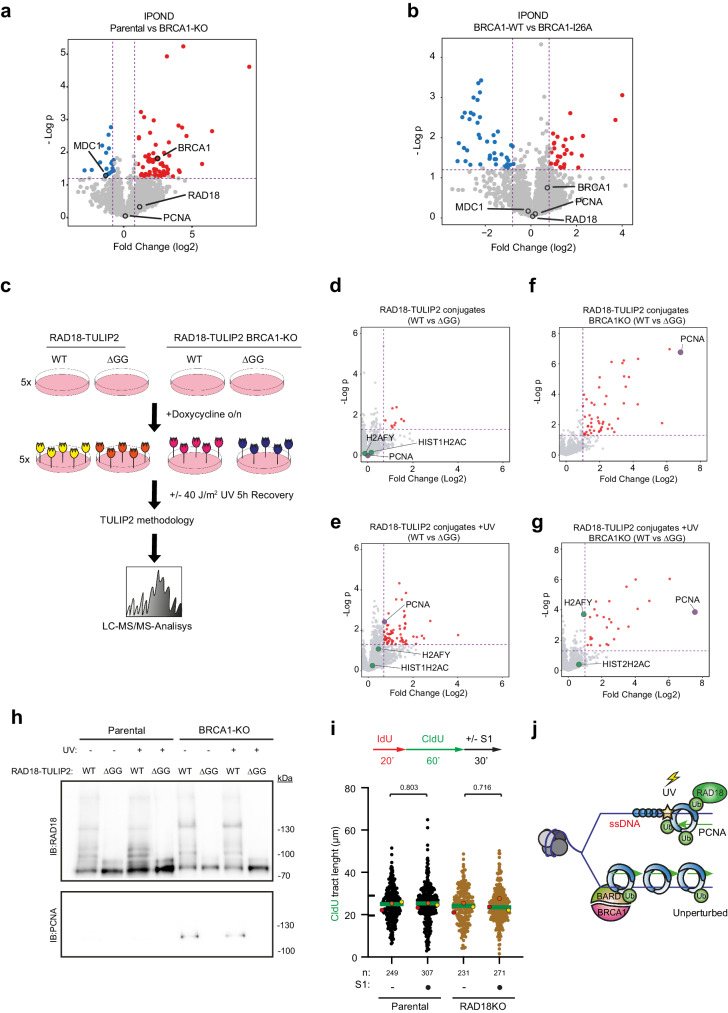
Fig. 4. RAD18 and BARD1 ubiquitinates PCNA in different pathways.
Volcano Plots depicting statistical differences in iPOND experiments comparing (a) Parental and BRCA1-KO cells or (b) BRCA1-WT or BRCA1-I26A rescued cells. P value corresponds to unpaired two-sided t-tests. Each dot represents a protein from Supplementary Data 4. c Schematic representation of RAD18-TULIP2 set up in Parental and BRCA1-KO cell lines. TULIP2 constructs were induced with 1 μg/μL doxycycline overnight. Cells were treated or not with 40 J/m2 of UV light and allowed to recover for 5 h prior to cell lysis, protein purification and identification by LC-MS/MS of the RAD18-TULIP2 conjugates. RAD18-WT constructs were compared with their correspondent ΔGG constructs for conjugates identification after MS analysis. Volcano plots of RAD18-TULIP2 conjugates without UV damage (d) and upon UV treatment (e) in Parental cell lines. Volcano plots depicting RAD18-TULIP2 conjugates without UV damage (f) and after UV damage (g) in BRCA1-KO cell lines. Histones are represented in green and PCNA is shown in purple. Each dot represents a protein from Supplementary Data 5 and proteins enriched over 0.8 fold (log2) and p > 0.05 are displayed in red. P value corresponds to unpaired two-sided t-tests. h Immunoblotting of RAD18-TULIP2 pull down samples with and without UV damage in Parental and BRCA1-KO cells. Antibodies used are indicated. This experiment was repeated three times with similar results. i Formation of ssDNA gaps in unperturbed conditions in Parental, and RAD18-KO cells. Top: Scheme of the IdU/CldU pulse-labeling protocol, followed by S1 nuclease treatment. Bottom: CldU tract lengths in the indicated cell lines with and without S1 nuclease treatment. Each dot represents one fiber and the green bar represents the median. n: indicates number of fibers measured from three biological independent experiments. p values were calculated using two-sided Mann-Whitney tests. Colored dots indicate the median of each independent experiment. j Proposed model for PCNA ubiquitination in two different scenarios. During unperturbed conditions BRCA1/BARD1 ubiquitinates PCNA at low levels to promote continuous DNA synthesis. Upon replication fork barrier-inducing lesions, such as UV irradiation, RAD18 ubiquitinates PCNA at higher levels. Source data are provided as a Source Data file. c It has been adapted and modified from Yalçin Z, Koot D, Bezstarosti K, Salas-Lloret D, Bleijerveld OB, Boersma V, Falcone M, González-Prieto R, Altelaar M, Demmers JAA, Jacobs JJL. Ubiquitinome Profiling Reveals in Vivo UBE2D3 Targets and Implicates UBE2D3 in Protein Quality Control. Mol Cell Proteomics. 2023 Jun;22(6):100548. 10.1016/j.mcpro.2023.100548. which was published under CC-BY license [https://creativecommons.org/licenses/by/4.0/].