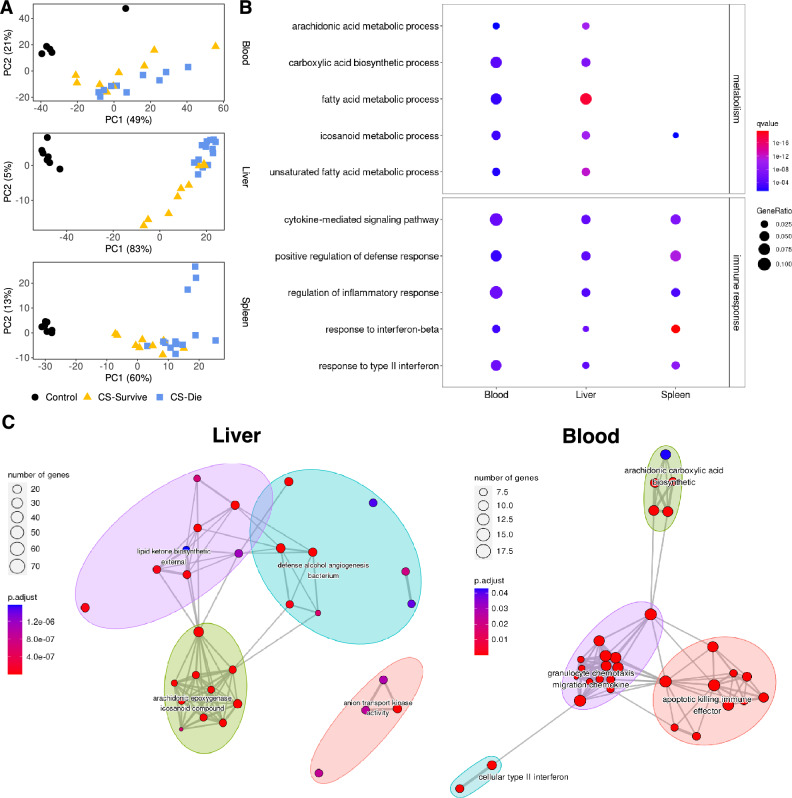
Figure 3.
Whole blood, liver and spleen transcriptomics comparing likely survivors and non-survivors of murine neonatal sepsis. (A) Unsupervised clustering of normalized RNA-seq data using principal component analyses of transcriptomic data collected from blood 1, liver (middle) and spleen (bottom); (n = 7 Control, 9 CS-Survive, 10 (blood) or 13 (liver, spleen) CS-Die; derived from 5 independent experiments). (B) Selected immune response and metabolic pathways from top enriched GO terms (by q-value) from differentially expressed genes in predicted survivors and non-survivors in the blood, liver, and spleen. (C) Network of clustered enriched GO biological process terms from differentially expressed genes in predicted survivor vs non-survivor mice in the liver and whole blood. The size of the node reflects the number of genes in the term, while the colour represents the adjusted p-value. Each functional cluster is represented by a different color. Edges connect terms that have shared genes (similarity > 0.2), and the length of the edge is inversely proportional to term similarity.