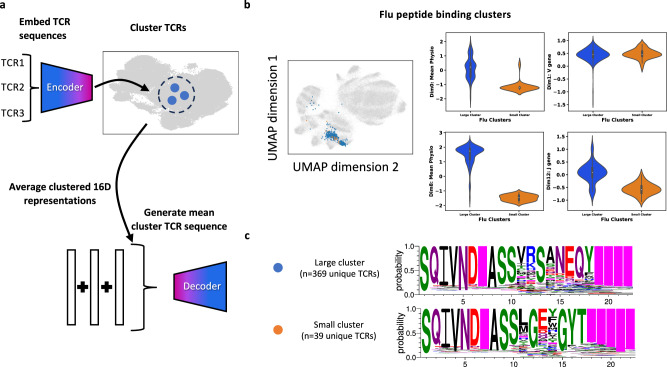
Fig. 5. TCR-VALID can be used to both identify TCR-Antigen binding modes and generate meta TCR sequences.
a Unique TCRs can be embedded and clustered in TCR-VALID latent space, their 16D representations can then be averaged to create meta representation of the TCR cluster. This meta representation can then be decoded to generate the meta-TCR sequence representative of the cluster. b TCRs from validated TCR-antigen pairs embedded into the learned latent landscape (left) and TCRs from the two largest flu clusters (Large cluster n = 369 unique TCRs and Small cluster n = 39 unique TCRs) embedded into learned latent landscape. Violin plots of latent dimensions of TCR-VALID model for two flu clusters above illustrating separation of clusters along Mean Insert Physicochemical properties and J gene usage but overlapping for V gene usage (right). White dot, median. Box edges, 25th and 75th quartiles. Whiskers, 1.5 × the IQR of the box edge. c TCR PWMs generated from the mean TCR-VALID representations of the two largest flu clusters, illustrating conserved CDR3 motif and gene usage. Source data for b are provided as a Source Data file.