Figure 1.
Method overview
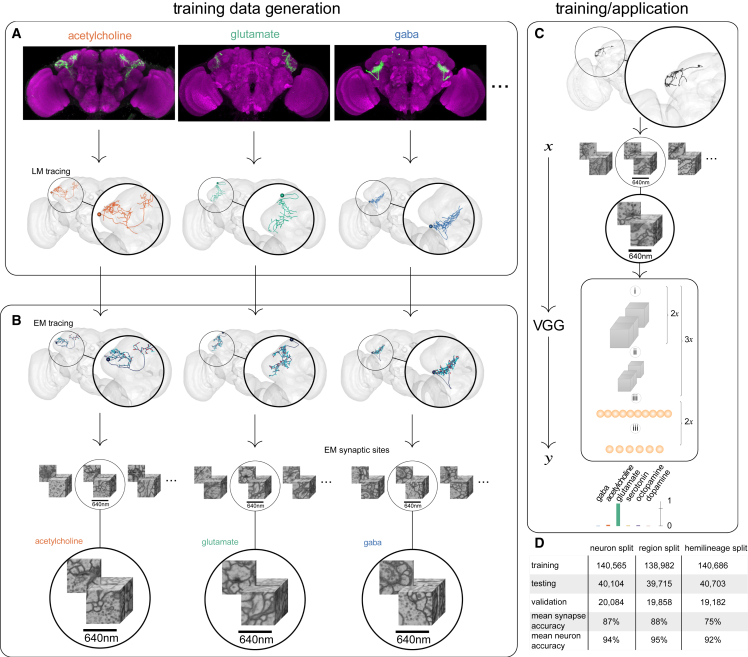
We assembled a dataset of neurons with known transmitter expression (see STAR Methods) in two D. melanogaster brain EM datasets (FAFB and HemiBrain) and retrieved corresponding synaptic locations.
(A) Typically, neurons had been genetically tagged to identify their transmitter identity and reconstruct their coarse morphology using light microscopy (Data S1).
(B) Light microscopy tracings of neurons are then matched to corresponding EM reconstructions with annotated synaptic locations, yielding a dataset of EM volumes of synaptic sites with known transmitter identity.
(C) We used the resulting pair (x, y) where x is a 3D EM volume of a synaptic site and y is the transmitter of that synaptic site (one of GABA, acetylcholine, glutamate, serotonin, octopamine, or dopamine) to train a 3D VGG-style deep neural network to assign a given synaptic site x to one of the six considered transmitters. We used the trained network to predict the transmitter identity of synapses from neurons with so far unknown transmitter identity. Panels i, ii, and iii denote convolution, down-sampling, and fully connected layers, respectively.
(D) Overview of our results on the FAFB dataset. Shown are the number of presynapses for training, testing, and validation as well as average synapse and neuron classification accuracy on the testing set for each data split.
See also Data S2.