Figure 2.
The accuracy of the trained classifier on a per-presynapse and per-neuron basis
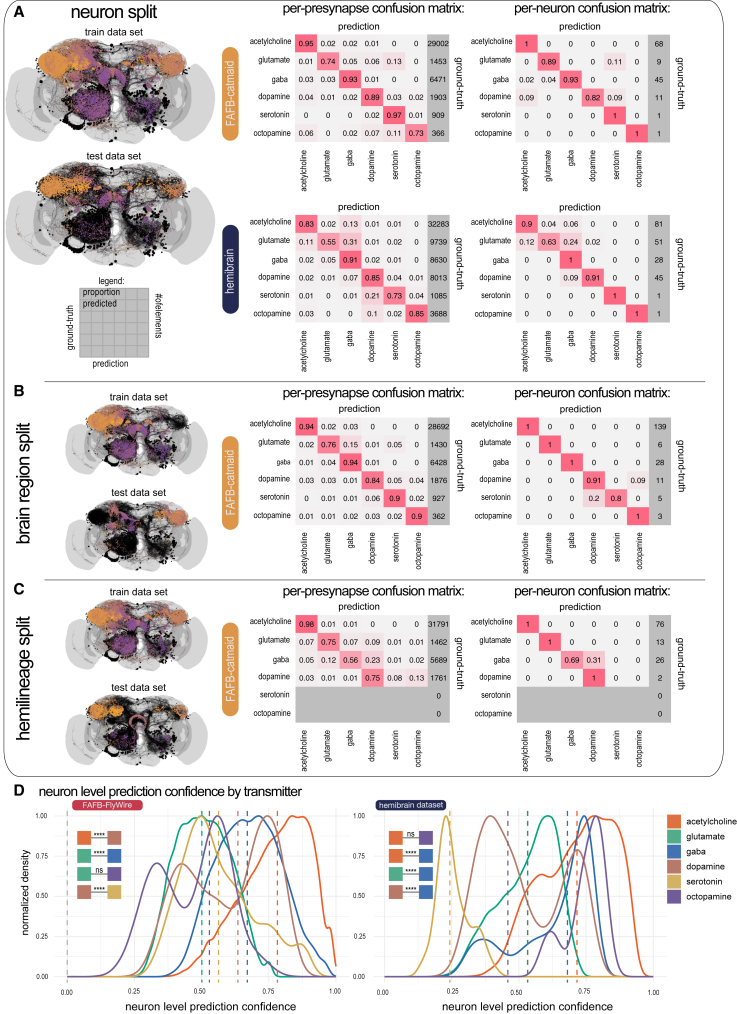
(A) Left: visualization of the training (upper) and testing (lower) data (split by entire neurons) that we used for the results in this manuscript. Presynapse locations are color coded according to their z-depth; anterior-posterior shown as purple-orange, neuron skeletons in black. Right: confusion matrices for the trained classifier on the testing data, shown per presynapse and as a majority vote per neuron, on datasets FAFB and HemiBrain. We considered only those neurons with more than 30 presynapses.
(B) Classification results on alternative training and testing data (split by brain regions) from FAFB.
(C) Same as (B) but split by hemilineage. It was not possible to generate a fully balanced split and as a result there are no serotonin and octopamine neurons in the testing set, as indicated by the grayed-out rows.
(D) The distribution of neuron-level confidence scores by transmitter, across our pool of central brain neurons in the FlyWire and HemiBrain datasets (FAFB-FlyWire, 136,927; HemiBrain, 24,666). Vertical dashed line, median value. Colored boxes with stars indicate statistical comparisons, Wilcoxon two-sample tests (n.s., not significant; ∗p ≤ 0.05; ∗∗∗∗p ≤ 0.00001).