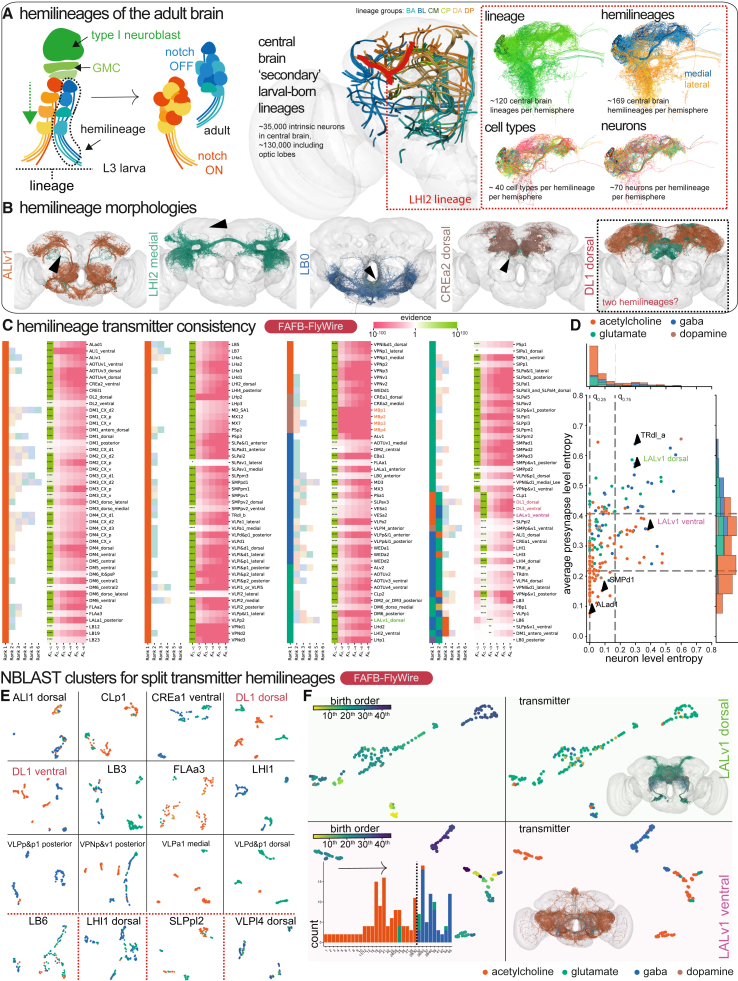
Figure 7.
Transmitter usage across all hemilineages in the central fly brain
(A) Left, the progression of a type I neuroblast from third-instar larva (L3) into the adult ganglion mother cell (GMC). Right, a breakdown of a single secondary lineage, “LHl2” into its constituent hemilineages (see STAR Methods).
(B) Example of homologous FAFB-FlyWire hemilineages on both sides of the brain, colored by neuron-level transmitter prediction. Black arrows point to one stray member of each hemilineage with a different neuron-level transmitter prediction, which is likely a first-born neuron with distinct morphology31,102 (Data S5). The dashed box indicates a hemilineage with potential split transmitter expression.
(C) Bayes factor analysis of hemilineage consistency. For each hemilineage (row), the right set of columns corresponds to the likelihood ratio of the hemilineage expressing that number of transmitters versus the likelihood of any other number (). Evidence strength: substantial ∗K ⩾ 101/2; good ∗∗K ⩾ 101; strong ∗∗∗K ⩾ 103/2; decisive ∗∗∗∗K ⩾ 102.103 The left set of columns indicates the frequency ranked transmitter predictions within the hemilineage, with ranks greater than the maximum likely number of transmitters shaded lighter. “LB0 posterior” did not have substantial evidence for any particular number of transmitters.
(D) Neuron level entropy (H(Nh)) versus average synapse level entropy (H(Sh)) for all predicted hemilineages with more than 10 neurons and more than 30 presynapses per neuron. Dashed lines indicate 25% and 75% percentiles: q25(H(Nh)) = 0.00, q25(H(Sh)) = 0.22, q75(H(Nh)) = 0.17, and q75(H(Sh)) = 0.41.
(E) NBLAST UMAP plots of selected hemilineages that exhibit some degree of predicted split transmitter usage. UMAPs are based on NBLAST morphological similarity scores between all possible pairs of neurons in each hemilineage. Points represent neurons, colored by neuron-level transmitter prediction. Black solid lines bound examples with a morphology-transmitter split, red dashed lines bound examples with no such clear divide. Red text label examples for which data annotation issues may explain split usage.
(F) “LALv1” has two hemilineages: dorsal (developmentally defined by Notch-ON) and ventral (Notch-OFF). NBLAST UMAPs for the two “LALv1” hemilineages (rows): dorsal (developmentally defined by Notch-ON) and ventral (Notch-OFF), colored by birth order (left) and neuron-level transmitter prediction (right). Bottom left, histogram of neuron-level transmitter prediction by birth order.102