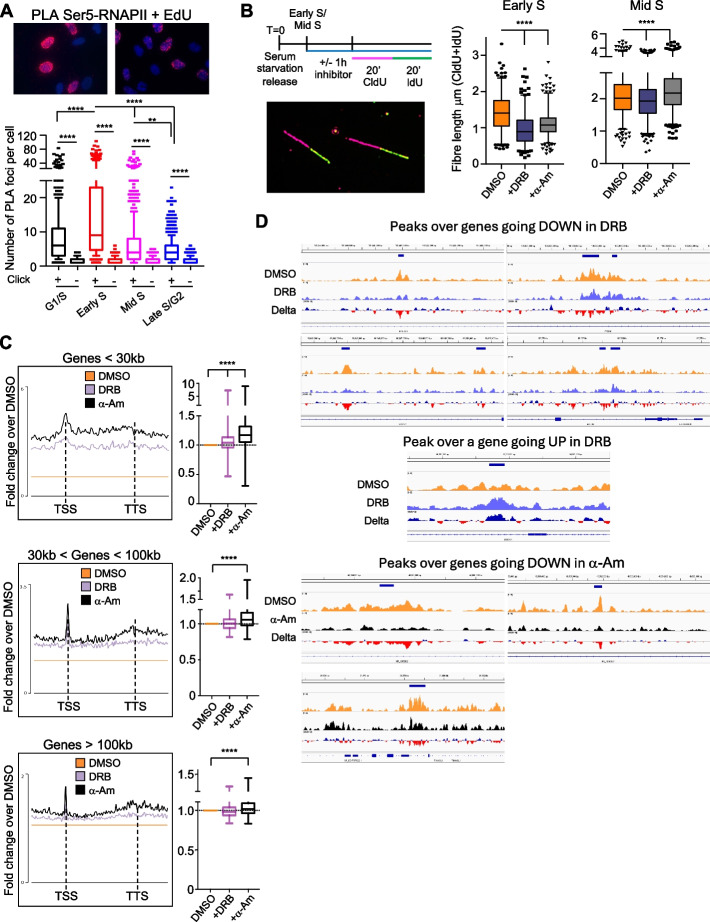
Fig. 3.
Transcription elongation affects DNA replication rates. A Proximity ligation assay (PLA) with cells treated with EdU and anti-Ser5-P-RNAPII antibody; representative images with DAPI staining in blue for nuclei and PLA foci in red; cells are synchronized as Fig. 1A and harvested at the indicated timepoints after 10 min incubation with EdU. PLA foci count in cells with 1 PLA foci (right panel), box-whisker plots 10–90 percentile with line at the median, Mann–Whitney t-test. n = 3, > 150 cells counted per timepoint in each repeat. B Schematic of the experiment with a representative image of DNA fibers; cells pretreated for 1 h with DRB (100 μM) or α-amanitin (α-Am, 3 μg/ml) before each timepoint as in Fig. 1A, followed by 20 min pulse with CldU and 20 min pulse of IdU; DNA fiber lengths measured using ImageJ; n = 3, > 400 fibers analyzed per condition; box whiskers plots with line at the median 2.5–97.5 percentile, Mann–Whitney t-test. C Average metagene profiles of BrdU-Seq from TSS to transcription termination site (TTS) for transcribed genes specifically replicated across in the first timepoint clustered by gene length, in cells treated for 1 h with DMSO, DRB (100 μM), or α-amanitin (α-Am, 3 μg/ml) before the Early S timepoint while being pulsed with BrdU, with quantifications of the BrdU-Seq signals from TSS to TTS in the DRB or α-amanitin samples as fold changes normalized to DMSO; paired nonparametric t-test. D IGV snapshot over BrdU peaks (highlighted by the blue bar) in genes replicated in the first timepoint that change expression levels following treatment with DRB or α-Am. BrdU-Seq profiles in DMSO (orange), α-Am (black), and DRB (blue) treated cells; delta represents the combination of the DRB or α-Am track with the DMSO one (DRB—DMSO or α-Am—DMSO), with blue signal indicating higher BrdU-Seq levels in DRB or α-Am compared to DMSO and red indicating higher BrdU-Seq levels in DMSO compared to the transcription inhibitors. * ≥ p-value < 0.05, ** ≥ p-value < 0.01, **** ≥ p-value < 0.0001