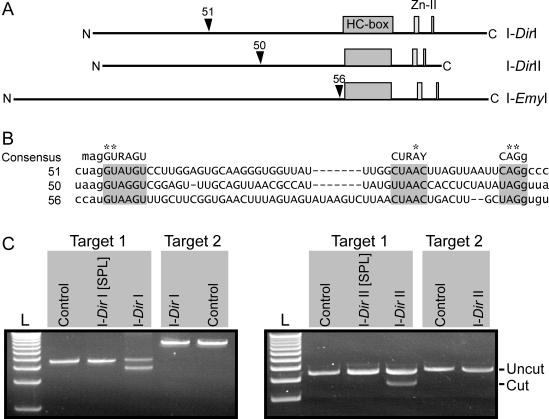
Figure 2.
Properties of Didymium His-Cys box HEGs. (A) The HEGs encoding the I-DirI and I-DirII homing endonucleases are interrupted by spliceosomal introns of 51 and 50 nt in size, respectively. A similar spliceosomal intron-like sequence has been identified in the I-EmyI HEG (6). The positioning of the HEG intervening sequences relative to the primary protein structure is shown (arrowheads), as are the N- and C-terminal ends. (B) Sequence alignment of spliceosomal introns (upper case letters) in nuclear HEGs (as described in A), with some flanking HEG sequences (lower case letters). The 51, 50 and 56 nt spliceosomal introns that interrupt the I-DirI, I-DirII and I-EmyI HEGs, respectively, share strong similarity to the mammalian spliceosomal intron consensus (on top) (31), including invariable positions (asterisks). Consensus nucleotides are A or C (M), A or G (R) and C or U (Y). (C) Endonucleolytic activity of homing endonucleases. Fused to the glutathione-S-transferase partner the homing endonucleases encoded by Intron 1 and Intron 2 were expressed with (I-DirI[SPL] and I-DirII[SPL], respectively) or without (I-DirI and I-DirII, respectively) interrupting spliceosomal introns. Affinity-purified fusion proteins were incubated with two different DNA targets under conditions that favor activity of the I-PpoI homing endonuclease (14). Target 1 contained the intron-lacking Didymium SSU rDNA, whereas Target 2 contained the Didymium SSU rDNA interrupted by the respective group I intron (i.e. Intron 1 or Intron 2). Endonuclease activity was recognized by cleavage of the target plasmid generating 3.83 and 0.77 kb fragments (0.77 kb band not shown). As a negative control, the DNA was incubated in the absence of proteins.