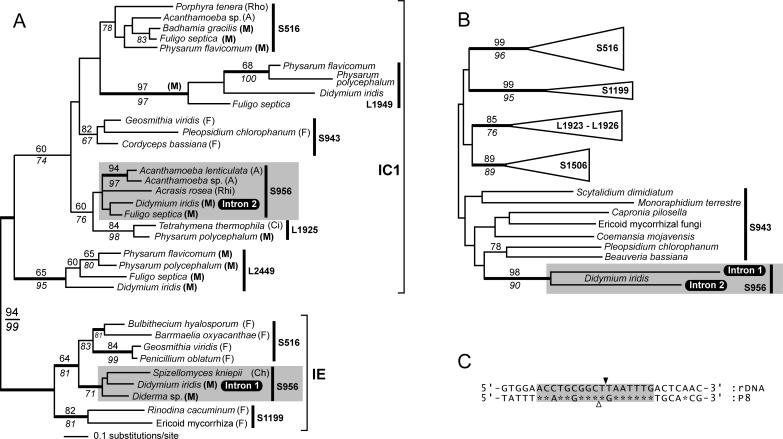
Figure 3.
Phylogeny of nuclear group I introns and homing endonucleases. (A) The intron tree was generated from a 131 nt dataset using the neighbor joining (NJ) method and the Jukes–Cantor (JC) evolutionary model. Bootstrap values above the branches were calculated using the NJ method (Kimura 2 model and 2000 replicates) and values below the branches are from a maximum parsimony (MP) analysis. Values >60% are shown. Support for branches was also tested using Bayesian analysis, and thicker branches indicate ≥95% posterior probability. Introns were sampled from fungi (F), Acanthamoeba (A), Myxomycota (M), Rhodophyta (Rho), Rhizopoda (Rhi), Ciliophora (Ci) and Chytridiomycota (Ch) (classification of organisms follows that of Margulis and Schwartz (32). Introns that belong to the group IC1 or group IE subclasses are indicated, and the site of intron insertion are shown (e.g. S516, S956 and L1925). The Didymium Intron 1 and Intron 2 belong to distantly related lineages of S956 introns (shaded fields). (B) Phylogeny of His-Cys box homing endonucleases. The tree is a summary of a minimum evolution (ME)-Poisson tree. Statistical support for branches was tested by ME-Poisson (2000 replicates) and NJ-JTT (500 replicates) bootstrap analyses, and the resulting numbers are shown above and below branches, respectively. We also tested the ME-tree by calculating Bayesian posterior probabilities using the JTT model. Supported branches in the ME tree are in agreement with a previous analysis (6), and show the clustering of homing endonucleases from homologous or neighboring (i.e. L1923–L1926) sites. The close phylogenetic relationship between the Didymium homing endonucleases is strongly supported by high bootstrap values (i.e. 98 and 90) and significant posterior probabilities. (C) Comparison of sequences flanking Intron 2 and sequences flanking the Intron 2 HEG. The rDNA intron insertion site (black arrowhead) and the putative HEG insertion site (open arrowhead) are shown. Asterisks indicate sequence identity, and a 17 nt region with high identity is indicated (shaded field). The present identity in sequences supports that the HEG invaded Intron 2 in a homing-like process, and that the event took place in the recent evolution of this element [‘recent’ on an intron evolutionary time-scale can refer to events that took place thousands, or even millions of years ago (33)].