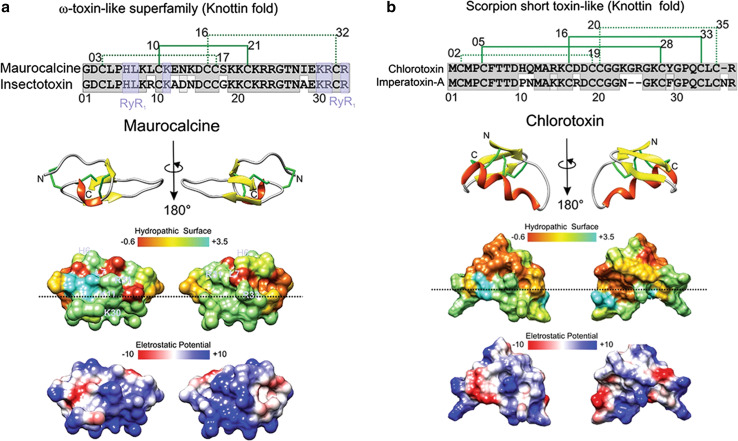
Fig. 4.
Structure and function of maurocalcine and chlorotoxin. a, b The primary, secondary and tridimensional surfaces of maurocalcine (GeneBank P60254; UniProt 1C6W [105]) and chlorotoxin (GeneBank P45639, UniProt 1CHL [106]) are shown, respectively. Gray conserved sequences; purple ryanodine receptor binding sequences (RyR1), based on [97]. The upper part of a corresponds to the alignment of maurocalcine (UniProtKB/Swiss-Prot: P60254.1) and insectotoxin I5A from the venom of the Mesobuthus eupeus scorpion (UniProtKB/Swiss-Prot sequence P15222.2). The upper part of b corresponds to the alignment of chlorotoxin (UniProtKB/Swiss-Prot sequence P45639.1) and imperatoxin-A from Pandinus imperator scorpion (UniProtKB/Swiss-Prot: P59868.1). The disulfide bridge pattern is shown in green. Lower portions of a and b indicate opposite surfaces of maurocalcine and chlorotoxin, with the distribution of hydrophobic and hydrophilic regions and charge distribution, based on [62, 104, 107]. Electrostatic surface colored according to Coulombic electrostatic potential, e = 4r, thresholds ± 5 kcal mol−1 e−1 at 298 K. Hydrophobicity surface colored following the Hessa and von Heijne hydropathic scale thresholds (dark orange most hydrophobic; white 0; aquamarine most hydrophilic) as described by Hessa et al. [99]. The dashed line shows peptide anisotropy, with highly hydrophobically charged residues above and positively charged sequences in the lower half. Secondary structure: beta-strand is depicted in yellow, alpha-helix in red, coil in pale gray and disulfide bonds are depicted in green. Molecular graphics and analyses for three-dimensional structures were performed with the UCSF Chimera package software [100]