Fig. 1.
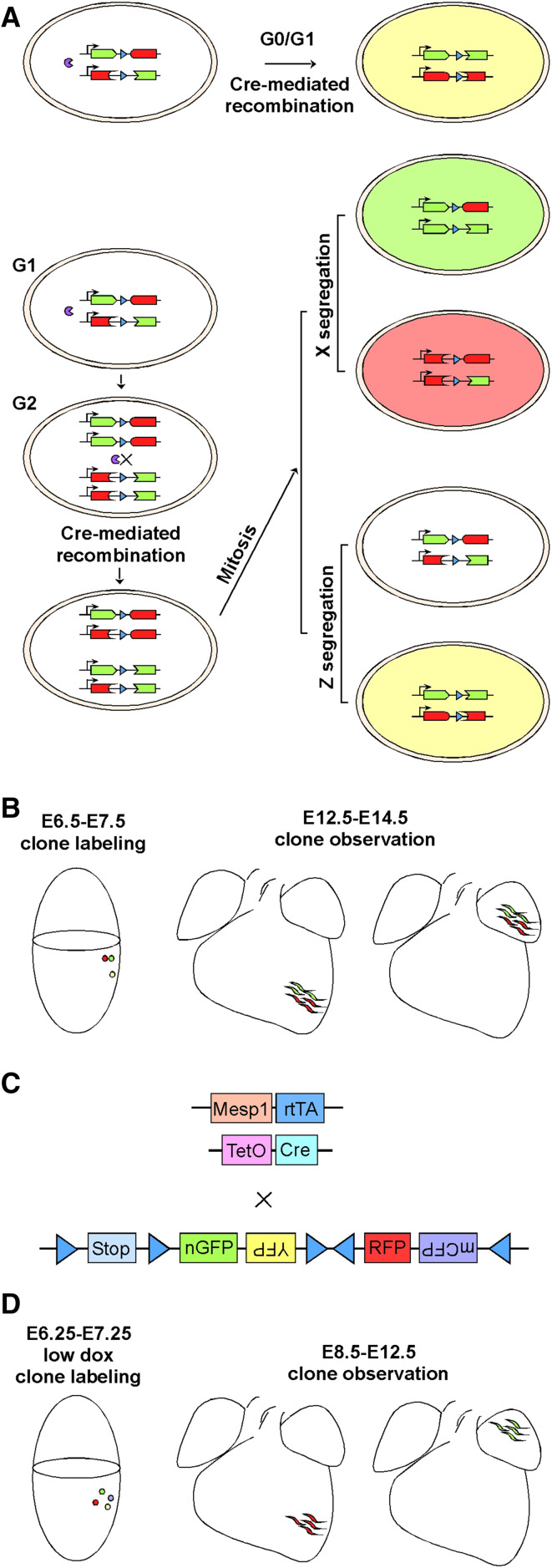
Tracking Mesp1-expressing cells at clonal level. a The principle of mosaic analysis with double markers (MADM). Before the expression of Cre recombinase, full-length GFP and tomato fluorescent proteins are not produced. Interchromosomal recombination is induced by Cre, restoring the expression of GFP and tomato. Recombination in G1 or postmitotic G0 cells leads to yellow color (expression of both GFP and tomato). Recombination in G2 cells and the subsequent mitosis leads to two scenarios. In X segregation, the two daughter cells are labeled either red or green. In Z segregation, the two daughter cells are labeled colorless or yellow. b Representative findings of Mesp1–Cre clonal labeling using the MADM system. c The strategy of clonal lineage tracking of Mesp1-expressing cells using the Rosa–Confetti reporter. d Representative findings in Mesp1-rtTA/TetO-Cre/Rosa–Confetti clonal labeling. A low dose of Dox is titrated and used to obtain unicolor embryos at the time of analysis