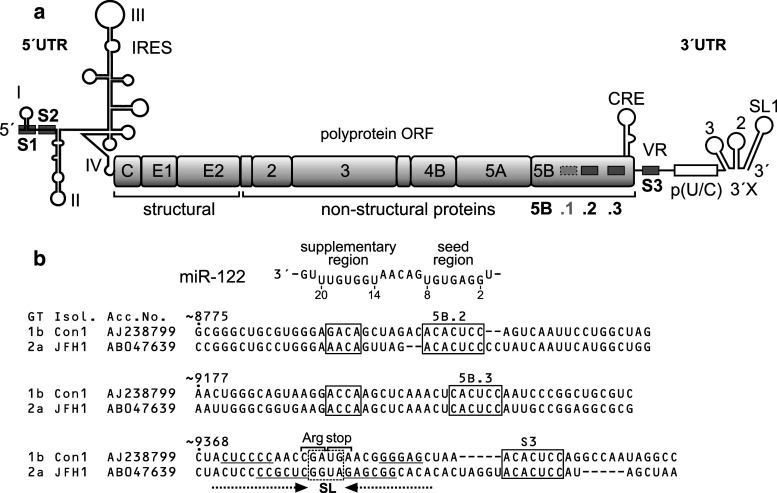
Fig. 1.
Conserved miR-122 target sites in the hepatitis C virus (HCV) RNA genome. a The HCV genome with the 5′- and 3′-untranslated regions (UTRs), the polyprotein open reading frame (ORF) and the gene segments encoding structural and non-structural (NS) proteins. In the 5′UTR, there are two conserved miR-122 target sites (S1 and S2, boxed) and the internal ribosome entry site (IRES). In the NS5B region (encoding the viral replicase) there are three miR-122 binding sites (numbered 5B.1, 2 and 3), of which sites 5B.2 and 5B.3 are highly conserved among isolates. One additional site (S3) is located in the variable region (VR) of the 3′UTR. The cis-acting replication element (CRE) in the NS5B region, the poly(U/C) tract and the highly conserved 3′X region in the 3′UTR are indicated. b miR-122 and conserved miR-122 target sites in the NS5B coding sequences and in the 3′UTR of two HCV isolates, Con1 (genotype 1b) and JFH-1 (genotype 2a). Upper part The miR-122 sequence with the seed sequence (nucleotides 2–8) and the potential supplementary binding region (involving nucleotides 14–20). Below Sequences of the conserved miR-122 target sites 5B.2, 5B.3 and S3 in the NS5B region and the 3′UTR in the Con1 (genotype 1b) and JFH-1 (genotype 2a) isolates. HCV genotype (GT) and NCBI nucleotide database accession numbers are given. Nucleotide numbers indicate numbering in the genotype 1b Con1 isolate. miR-122 target sequences are boxed. Alignment of 106 selected isolates and miR-122 seed and potential supplementary binding sites in the NS5B region are shown in Supplementary Figures S1, S2 and S5