Fig. 7.
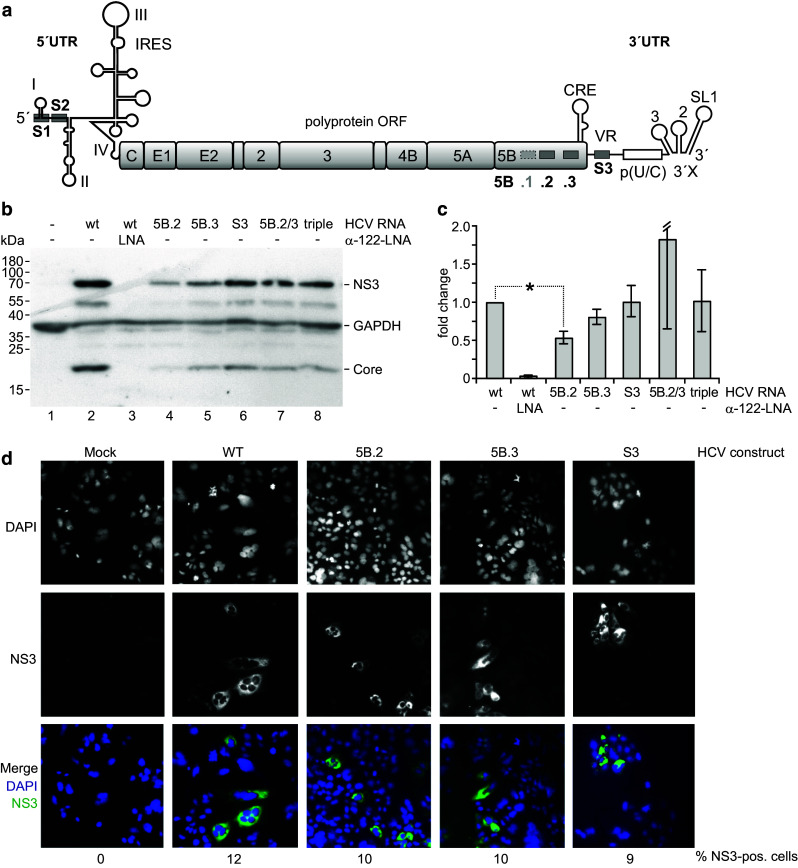
Effects of mutations in the miR-122 binding sites in the NS5B coding region and 3′UTR on HCV overall replication. a The HCV full-length genome with the miR-122 binding sites S1 and S2 in the 5′UTR, 5B.1, .2 and .3 in the NS5B coding region and S3 in the 3′UTR. Site 5b.1 was disabled in all constructs used here since this site is conserved only in a minor fraction of HCV isolates. b–d Cells were transfected with the wild-type HCV RNA or mutated variants as indicated. Cells were harvested 3 days after transfection, and aliquots of the cells extracts were used for analysis of protein expression (NS3 and Core) (b) and HCV plus strand genome abundancy by qPCR (c). d Shows expression of NS3 after 2 days by immune fluorescence. Nuclear staining by DAPI and NS3 staining each are shown in b/w, while the merge is shown in color (blue DAPI, green NS3). “5B.2/3” labels the double mutant with sites 5B.2 and 5B.3 mutated, “triple” the triple mutant with sites 5B.2, 5B.3 and S3 mutated in combination. The counted % of cells expressing NS3 is indicated below
