Fig. 3.
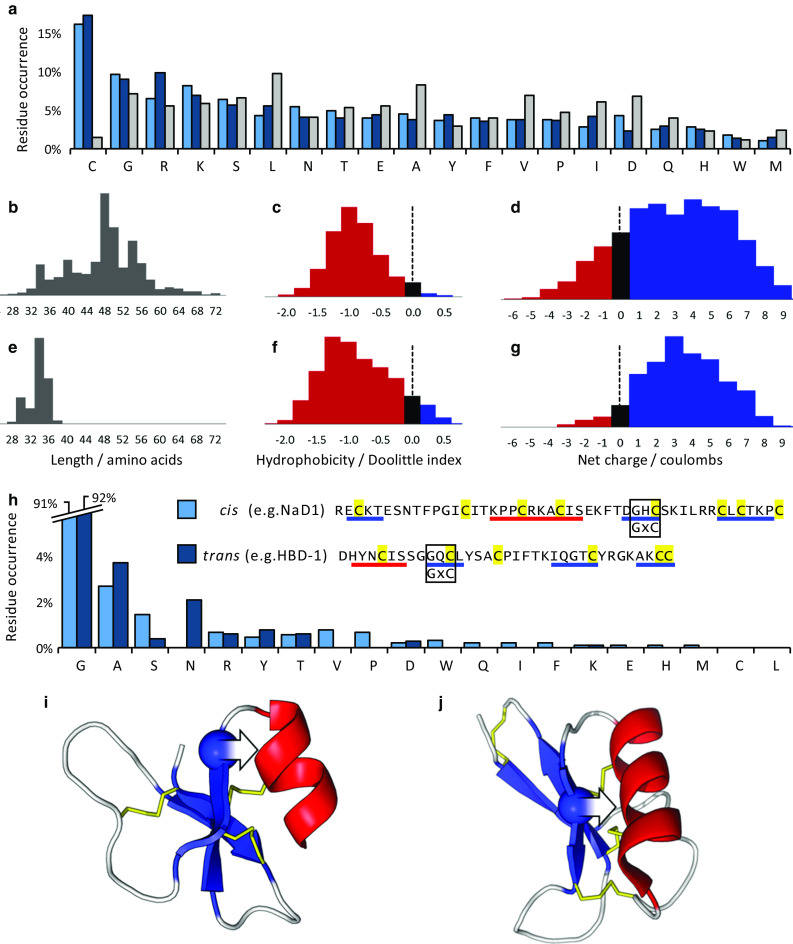
Amino acid sequence properties of cis- and trans-defensins. a Average amino acid residue occurrence for the cis-defensins (light blue), trans-defensins (dark blue) and whole Uniprot database (grey). Distributions of length, hydrophobicity and charge for b–d 1820 cis-defensins and e–g 894 trans-defensins. The common GxC motif occurs in both cis-defensins (e.g. NaD1) and trans-defensins (e.g. HBD-1). h Residue bias in the first position of the GxC motif in the cis-defensins (excluding S-locus and spiderines, which have an additional disulphide at this location) and the trans-defensins (excluding α- and θ-defensins, which lack an α-helix and so are unconstrained at this location). In both i cis-defensins (PDB:1MR4) and j trans-defensins (PDB:1IJV), the glycine (sphere) is oriented such that a non-hydrogen R-group on any other amino acid in this position (arrow) would clash with the α-helix. β-Strands in blue, α-helices in red, disulphide bonds in yellow
