Abstract
The genome integrity of all organisms is constantly threatened by replication errors and DNA damage arising from endogenous and exogenous sources. Such base pair anomalies must be accurately repaired to prevent mutagenesis and/or lethality. Thus, it is not surprising that cells have evolved multiple and partially overlapping DNA repair pathways to correct specific types of DNA errors and lesions. Great progress in unraveling these repair mechanisms at the molecular level has been made by several talented researchers, among them Tomas Lindahl, Aziz Sancar, and Paul Modrich, all three Nobel laureates in Chemistry for 2015. Much of this knowledge comes from studies performed in bacteria, yeast, and mammals and has impacted research in plant systems. Two plant features should be mentioned. Plants differ from higher eukaryotes in that they lack a reserve germline and cannot avoid environmental stresses. Therefore, plants have evolved different strategies to sustain genome fidelity through generations and continuous exposure to genotoxic stresses. These strategies include the presence of unique or multiple paralogous genes with partially overlapping DNA repair activities. Yet, in spite (or because) of these differences, plants, especially Arabidopsis thaliana, can be used as a model organism for functional studies. Some advantages of this model system are worth mentioning: short life cycle, availability of both homozygous and heterozygous lines for many genes, plant transformation techniques, tissue culture methods and reporter systems for gene expression and function studies. Here, I provide a current understanding of DNA repair genes in plants, with a special focus on A. thaliana. It is expected that this review will be a valuable resource for future functional studies in the DNA repair field, both in plants and animals.
Keywords: DNA repair, Photolyases, BER, NER, MMR, HR, NHEJ
Introduction
All organisms are constantly exposed to environmental stresses as well as genotoxic products from endogenous metabolic processes that induce, either directly or indirectly, DNA damage [1]. Another source of DNA damage includes base–base mismatches produced by the incorporation of a non-complementary Watson–Crick base and unpaired nucleotides caused by misalignment of the two DNA strands, both generated during DNA synthesis. These DNA biosynthetic errors differ from typical DNA damage in that they involve undamaged bases and exist only transiently provided that the DNA remains double-stranded [2]. To counteract these genome integrity threats, all organisms have evolved the DNA damage response (DDR) [3, 4]. The DDR is a pathway that transduces the DNA damage signal into activation of various pathways that leads to DNA repair, cell cycle checkpoint and programmed cell death. Plant DDR also includes endoreduplication. Endoreduplication involves replication of the nuclear DNA without cytokinesis [5]. Here, I only focus on DNA repair mechanisms in mammals and plants. Plants are distinctly different from mammals in that they lack a reserved germ line and are sessile. Gametes arise from meristem cells that have been exposed to the continuous effect of environmental and endogenous mutagens and have divided many times. Thus, plants have evolved particularly efficient DNA repair mechanisms. Advancements in this field have been provided by the Arabidopsis genome sequence [6]. Plants have orthologs of most of the genes involved in mammalian DNA repair pathways [7–9]. However, several interesting features may be noted: the presence of unique genes, the presence of multiple gene copies and the absence of well-characterized genes in plant genomes [9, 10]. Such differences suggest plant-specific DNA repair pathways. Provided here is a review of major findings over the last decade related to mammal and plant DNA repair mechanisms, with a special focus on DNA repair genes from A. thaliana.
Photoreactivation
Photoreactivation is the direct reversal of major lesions induced by ultraviolet (UV) light on two adjacent pyrimidine bases, such as cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidones (6-4 photoproducts or 6-4 PPs). The history of the discovery of this repair system has been recently reviewed [11]. Photoreactivation relies on single light-activated flavoenzymes called photolyases. Photolyases belong to the photolyase/cryptochrome family and have been classified into three related subfamilies known as class I CPD photolyase, class II CPD photolyase, and 6-4 photolyase, based on their sequence homology and substrate specificity [12–14]. Multiple sequence alignment analysis of members from the three subfamilies indicates a highly functional conservation of a photolyase-homologous region (PHR) domain that binds the chromophore FAD (flavin adenine dinucleotide) [15, 16]. Class I CPD photolyases have been reported in diverse microbial organisms such as archaea, eubacteria, and fungi; class II CPD photolyases have been found not only in archaea, eubacteria, some viruses, but also in higher organisms such as animals and plants; and 6-4 photolyases have been identified in cyanobacteria and certain eukarya [15, 17]. Humans and other placental animals have lost photolyase activities. All photolyases use the energy of visible or blue light (λ = 320–500 nm) for the catalytic cleavage of the photoproducts and contain the fully reduced FADH− as the catalytically active cofactor [13, 14]. All photolyases characterized to date also contain a second chromophore, such as methenyltetrahydrofolate found in the majority of the enzymes or 7,8-didemethyl-8-hydroxy-5-deazariboflavin, FAD or FMN (flavin mononucleotide) in others that functions as a photoantenna to enhance light absorption [14, 18–20]. Upon blue light absorption, the excitation energy is efficiently transferred from the photoantenna to FADH− by Förster resonance energy. The resulting FADH−* donates an electron to the DNA lesion with the generation of two canonical pyrimidines [12, 14, 19–21].
Photoreactivation in plants
Arabidopsis thaliana contains two specific class II CPD photolyases [PHR1, also named UVR2 (At1g12370) and PHR2 (At2g47590)] and one 6-4 photolyase [UVR3 (At3g15620)]. The FAD-binding domains of plant PHR2 proteins are smaller and less conserved than in the other subfamilies [15]. Several articles provide a summary of the molecular cloning, the tissue-specific and light-dependent regulation and the characterization of mutants of CPD photolyase and 6-4 photolyase genes from various plant species (e.g., A. thaliana, Oriza sativa, Sinapis alba) [22–24]. Recent evidence suggests that AtPHR1 expression induced by UV-B is primarily regulated by the UVR8 (UV RESISTANCE LOCUS8)-dependent pathway but is also mediated by a UVR8-independent pathway [25]. The latter one was found to be correlated with UV-B-induced CPD levels [25].
The crystal structure of a UVR2 homolog has recently been determined in rice and compared with prokaryotic class I CPD photolyases [26]. Results indicate that both enzymes recognize CPD damage in similar active sites but create a different conformational distortion in the DNA duplex [26]. The crystallographic structure and molecular mechanism of A. thaliana 6-4 photolyase have also been determined [27, 28]. After light excitation, there is initially a forward electron transfer followed by a cyclic proton transfer involving an active-site histidine residue [28]. This proton transfer is the determinant step in the repair and determines the overall quantum yield (Ф = number of dimers repaired per number of photons absorbed) [28]. For comparison, the reported Ф for CPD and 6-4PP repair are 0.82 and 0.1, respectively, and the whole catalysis reaction takes less than a nanosecond for CPD repair or tens of nanoseconds for 6-4PP repair [29]. The complete photocycle in real-time from the initial 6-4 PP to several intermediates and subsequent conversion to two thymine bases catalyzed by the A. thaliana (6-4) photolyase has been recently well reviewed [21, 29].
Base excision repair
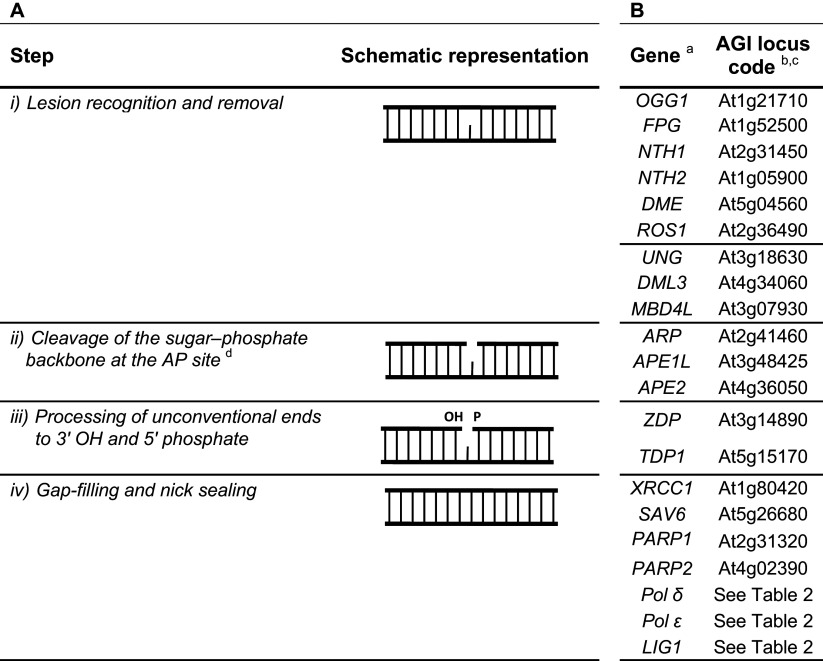
Base excision repair (BER) recognizes and repairs lesions such as deaminated, oxidized and alkylated bases, abasic (apurinic and/or apyrimidinic, AP) sites and single-strand breaks (SSBs). BER occurs in several steps (Table 1A; see references [30–32] for recent reviews in mammalian BER). (1) Lesion recognition and removal by DNA glycosylases. DNA glycosylases hydrolyze the N-glycosidic bond of the damaged base, leaving an AP site. Different DNA glycosylases exist, each one with broad or narrow substrate specificities. Enzymes can be classified as monofunctional or bifunctional. Monofunctional glycosylases perform base excision only, whereas bifunctional glycosylases also have lyase activity. Biochemical mechanisms of DNA glycosylases have been the subject of recent reviews [33, 34]. (2) Cleavage of the sugar–phosphate backbone at the AP site. Cleavage is catalyzed either by an AP endonuclease or by the intrinsic AP lyase activity of a bifunctional DNA glycosylase. AP endonucleases and lyases generate different types of DNA termini. AP endonucleases release a 3′ OH and a 5′ deoxyribose–phosphate moiety (5′ dRP), while AP lyases release a 3′ phospho a,β-unsaturated aldehyde (3′ PUA) or 3′ phosphate by β- or βδ-elimination reactions, respectively, and a 5′ phosphate. (3) Processing of unconventional ends to conventional 3′ OH and 5′ phosphate moieties. This step involves different enzymes depending on whether the modified terminus is processed from the 5′ or 3′ end. The intrinsic dRP lyase activity of DNA polymerase β removes the 5′ dRP; the 3′ phosphodiesterase activity of the AP endonuclease eliminates the 3′ PUA, whereas polynucleotide kinase 3′ phosphatase (PNKP) processes the 3′ phosphate moiety. Additional SSB end cleaning enzymes are aprataxin and tyrosyl DNA phosphodiesterases (TDP) that process abortive ligation or topoisomerase reactions, respectively [35]. (4) Gap-filling and nick sealing. The short-patch or single nucleotide (SN) BER involves the incorporation of only a single nucleotide by the DNA polymerase β. The long-patch (LP) BER requires DNA synthesis mediated by DNA polymerases δ and ε and assisted by proliferating cell nuclear antigen (PCNA). The newly synthesized DNA displaces the downstream 5′ DNA end to form a flap structure containing 2–13 nucleotides. The displaced strand is then processed by a flap endonuclease (FEN). The choice of the pathway depends on the specificity of the DNA glycosylase and the proliferation status of the cell. Recent findings suggest that DNA polymerase λ can substitute for DNA polymerase β in filling single-nucleotide gaps and can perform gap filling in long-patch BER [36]. Poly ADP-ribose polymerase 1 (PARP1) and X-ray cross-complementation group 1 (XRCC1) also have roles in BER. PARP1 protects SSBs from the formation of more deleterious double-strand breaks (DSBs) [35] and XRCC1 is involved in the organization of BER-proficient multiprotein complexes [37]. Finally, the nick is sealed by a DNA ligase.
Table 1.
Overview of steps (A) and A. thaliana required factors (B) involved in BER
aGenes encoding bifunctional glycosylases are listed first, followed by monofunctional glycosylases
b AGI Arabidopsis genome initiative
cAGI locus code for components involved in DNA metabolic pathways is shown in Table 2
dBifunctional DNA glycosylases with associated β-elimination activity are not detailed
BER in plants
Most of the BER proteins found in animal cells have been described in plants [38, 39]. Tables 1B and 2 show an overview of described factors involved in BER mechanism in A. thaliana. Plants contain several DNA glycosylases: 8-oxoguanine DNA glycosylase (OGG1), formamidopyrimidine DNA glycosylase (FPG), NTH, and uracil DNA glycosylase (UNG) [38–44]. The enzymes OGG1 and FPG recognize oxidized purines; NTH, oxidized pyrimidines and UNG, U:G mismatches. Both NTH homologs (NTH1 and NTH2) were co-localized to nucleoids within Arabidopsis chloroplasts, thus indicating the existence of a BER pathway in these organelles to deal with photo-oxidative stress [44]. Plant genomes, in contrast to mammals, also encode specific 5-methyl-cytosine (5-meC) glycosylases that are involved in DNA demethylation [DEMETER (DME) and Repressor of Silencing 1 (ROS1)] [45–48]. These proteins show preference for 5-meC as substrate but can also process T:G mispairs. It has been reported that ROS1 has a very low turnover [46], with the recognition of pyrimidine modifications being the rate-limiting step [49]. An additional activity against T:G mispairs is displayed by MBD4L (methyl binding 4 DNA glycosylase-like) [50]. This enzyme was demonstrated to act on both U:G and T:G mispairs and to excise both U and T more efficiently at a CpG context. Roles of OGG1, ROS1 and MBDL4 in Arabidopsis tolerance to oxidative DNA damage have also been described using mutant or overexpression plants [51–53]. Other downstream proteins of the BER pathway have been characterized in plants. The Arabidopsis genome contains three genes encoding AP endonuclease-like proteins: APE1L (abasic endonuclease-1-like), APE2 (abasic endonuclease 2) and ARP (abasic endonuclease-redox protein) [54]. All three enzymes were found to exhibit AP endonuclease activity in vitro [55], with ARP representing the major activity in Arabidopsis cell extracts [56]. Based on the available DNA sequence data, a wheat homolog of AtAPE1L has been cloned and characterized [57]. Orthologues of human PNKP (named AtZDP), TDP1, XRCC1 and DNA ligase I were also functionally characterized in Arabidopsis [42, 55, 56, 58–61]. Finally, it has been demonstrated that AP sites may be processed in plants through both SN- and LP-BER [41] and that DNA ligase I restores the continuity of the repaired DNA strand during both SN- and LP-BER [56]. Recent studies identified and characterized a gene named SAV6 (shade avoidance mutant), a homolog of human FEN1 [62]. Compared to the animal FEN1, SAV6 shows both flap and gap endonuclease activities, but lacks exonuclease activity.
Table 2.
Components involved in DNA metabolic pathways in A. thaliana
| Gene | EXO1 | PCNA | POL δ | POL ε | RFC | LIG1 |
|---|---|---|---|---|---|---|
| AGI locus code | At1g29630 |
At1g07370 At2g29570 |
At1g09815 At2g42120 At5g63960 |
At1g08260 At2g27120 At5g22110 |
At1g21690 At1g63160 At1g77470 At5g22010 At5g27740 |
At1g08130 At1g49250 |
Nucleotide excision repair
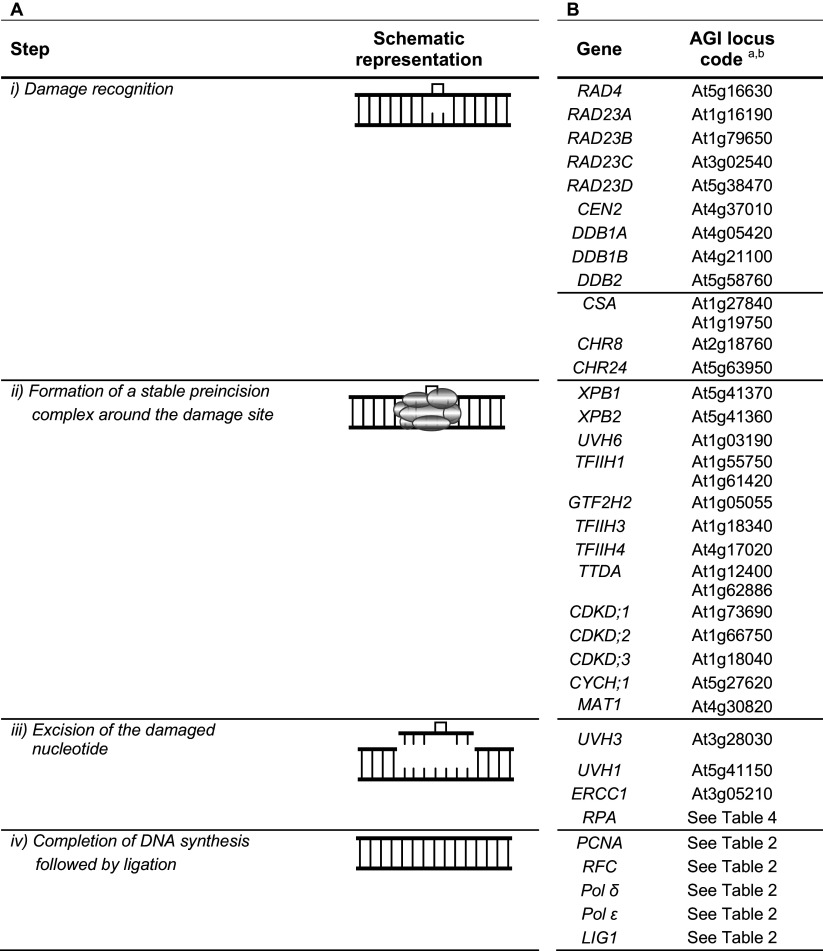
Nucleotide excision repair (NER) is responsible for the processing of bulky helix-distorting damage, such as CPDs and 6-4PPs induced by UV irradiation. Deficiencies in NER are associated with several human autosomal recessive disorders, namely xeroderma pigmentosum (XP), cockayne syndrome (CS) and trichothiodistrophy (TTD) among others. XP, CS and TTD have provided the names of some of the genes involved in the pathway (XPA through XPG, XPV, CSA, CSB and TTDA). NER is a multistep process which comprises four steps (Table 3A). (1) Damage recognition. Two different modes of damage recognition coexist: global genome NER (GG-NER) and transcription-coupled NER (TC-NER). GG-NER detects damage occurring throughout the genome and is specifically initiated by a heterotrimeric XPC-HR23B-CEN2 complex (XPC and HR23A/HR23B are the human homologs of yeast and plant RAD4 and RAD23 proteins, respectively), with the assistance in some cases of DDB (damaged DNA-binding) protein complex. DDB, a heterodimeric complex comprising DDB1 and DDB2 subunits, specifically binds CPDs and 6-4PPs with moderate or high affinity, respectively, and stimulates significantly XPC binding to UV-damaged lesions. GG-NER in mammalian cells has been covered in recent reviews [63, 64]. TC-NER is responsible for the recognition of lesions in the transcribed strand of active genes and is initiated by RNA polymerase stalled at a lesion, with the help of specific factors CSA, CSB, and XPA binding protein 2 (XAB2) whose function is not clear. Many aspects of mammalian TC-NER can be found in some recent reviews [64–66]. (2) Formation of a stable preincision complex around the damage site. After DNA recognition, GG-NER and TC-NER converge into the same pathway. The transcription factor II H (TFIIH) is recruited to the lesion. TFIIH is a multiprotein complex composed of ten subunits arranged in two functional subcomplexes: the core subcomplex containing the DNA helicase XPB, p62, p52, p44, p34 and p8 (TTDA) and the CAK [cyclin dependent kinase (CDK)-activating kinase] subcomplex containing CDK7, cyclin H (CYCH) and the assembly factor ménage-à-trois-1 (MAT1); both subcomplexes are bridged by the DNA helicase XPD [67, 68]. The result of the TFIIH activity is the partial unwinding of the DNA duplex that leads to the recruitment of XPA, RPA (replication protein A), and the endonuclease XPG. Then, XPA interacts and recruits the second endonuclease, ERCC1 (excision repair cross complementing 1)–XPF. (3) Excision of the damaged nucleotide. The damaged nucleotide is removed by dual incisions 5′ and 3′ to the lesion by endonucleases XPF and XPG, respectively, generating a 24–32 oligonucleotide single-strand fragment containing the damaged site. There is a defined order of the two incisions. The 5′ incision by the heterodimer ERCC1–XPF is performed first; repair synthesis is initiated followed by the 3′ incision by XPG [69]. (4) Completion of DNA synthesis followed by ligation. The repair synthesis is carried out by DNA polymerases δ, ε, or κ depending on chromatin accessibility of the damaged site [70]. NER is completed by nick sealing by DNA ligase I or IIIα.
Table 3.
Overview of steps (A) and A. thaliana required factors (B) involved in NER
NER in plants
This pathway has been primarily studied in Arabidopsis and rice [7]. In Arabidopsis, four genes, namely RAD23, DDB1, CSA and XPB have been duplicated (Table 3B) [7, 71]. Interestingly, RAD23 was also found to be duplicated in rice, Populus and Sorghum and many times in Arabidopsis [7, 71]. Although Arabidopsis mutants affecting the expression of individual RAD23 genes exhibited pleiotropic developmental defects (rad23b) or no obvious phenotype (rad23a, rad23c, and rad23d), higher-order mutant combinations indicated that the RAD23 family is essential in Arabidopsis [72]. The two DDB1 homologs, DDB1A and DDB1B, were expressed throughout plant development with DDB1B showing the lowest levels [73, 74] but playing a crucial role because ddb1b null alleles appear lethal ([73] and references therein). Overlapping expression patterns were also observed for the two CSA homologs. Both homologs share significant sequence identity and likely form heterotetramers in plants [75]. Decreased levels of several of these genes, namely DDB1A, DDB1B, CSA or XPB1 affected UV plant tolerance [75–79]. A difference in sensitivity upon exposure of UV radiation was also observed for plants defective in DDB2, UVH6 (defective in the human XPD homolog) or CSB homologs (CHR8 and CHR24) [76, 77, 80–82]. It should be mentioned that some plant homologs of the human XP genes were initially named after the UV hypersensitive (uvh) phenotype of the mutants. On the other hand, overexpression of DDB1A conferred increased resistance to UV-induced DNA damage [78].
The interaction of XPC-CEN2 (AtRAD4-AtCEN2) and validation of some in silico predicted TFIIH subunit transcripts were also reported in plants [7, 83–87]. Human TFIIH components p62, p44, p34 and p52 correspond to TFIIH1, TFIIH2 (AtGTF2H2), TFIIH3 and TFIIH4 in A. thaliana, while CDK7 is encoded by three CDKDs (Table 3B). Homologs of the human RPA complex, heterotrimer composed of three associated subunits RPA1, RPA2 and RPA3, were also functionally characterized in Arabidopsis [88]. However, in contrast to other eukaryotes, multiple genes encode the RPA subunits in A. thaliana (Table 4) [88, 89]. A phylogenetic tree based on RPA1 protein sequences revealed three evolutionary groups [90]. Group A is composed of RPA1A; group B, of RPA1B and RPA1D and group C, of RPA1C and RPA1E proteins. These authors propose that group C includes proteins involved in DNA damage repair [90]. No apparent XPA homolog exists in plants [10].
Table 4.
RPA proteins in A. thaliana
| Gene | RPA1 | RPA2 | RPA3 |
|---|---|---|---|
| AGI locus code |
At2g06510 (A) At5g08020 (B) At5g45400 (C) At5g61000 (D) At4g19130 (E) |
At2g24490 (A) At3g02920 (B) |
At3g52630 (A) At4g18590 (B) |
Mismatch repair
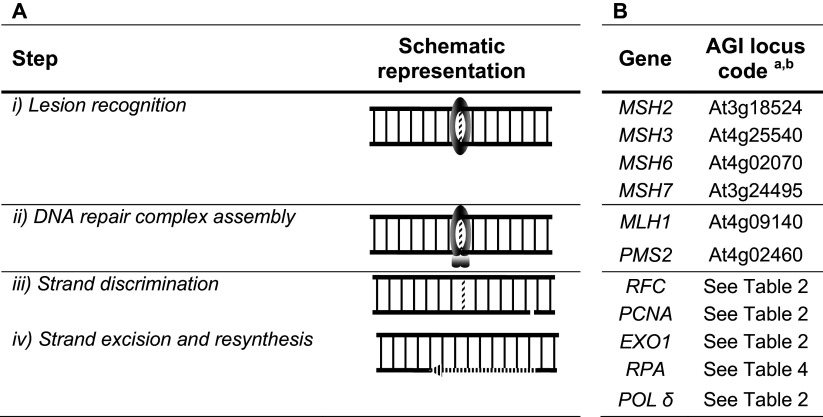
The mismatch repair (MMR) system is best known for its role in the recognition and correction of single base–base mismatches and unpaired nucleotides that arise through replication errors, deamination of 5-methylcytosine, and recombination between divergent sequences. Eukaryotic MMR has been studied in great detail and has been covered in several recent reviews [2, 91–94]. Also, an overview of the history of eukaryotic MMR has been recently reported [95]. The pathway involves several steps (Table 5A). (1) Lesion recognition by MutS proteins. In eukaryotes, MutS proteins function as heterodimers composed of related, but distinct MutS homologs (MSH) subunits. To date, eight MSH subunits were discovered [96], of which MSH2, MSH3 and MSH6 are involved in nuclear MMR in human cells. These MSHs subunits assemble as MSH2-MSH6 (MutSα) and MSH2-MSH3 (MutSβ). MutSα recognizes base–base mismatches and short insertion/deletion loops (IDLs) while MutSβ mediates the repair of IDLs up to 16 nucleotides [97]. The molecular basis of substrate specificities has been investigated. A series of crystal structures of human MutSα in complex with different DNA substrates showed an extensive interaction of a conserved phenylalanine residue in the MSH6 subunit with the DNA mispair [98]. MSH3 lacks the phenylalanine residue that makes protein-mispair contact. Instead, MutSβ binds the sugar–phosphate backbone of the IDL [99]. DNA is severely bent and the unpaired bases are flipped out and solvent exposed as revealed by the crystal structure of human MutSβ complexed with DNA containing IDLs of varying size [99]. In addition to crystal reports, abilities of MutSα and MutSβ to process base–base mismatches and IDLs were explored. The results indicate that MutSα and MutSβ differ in DNA substrate recognition and that the MutSβ–IDL interaction is greatly stimulated by an excess amount of MutSα [97]. Besides DNA-binding activity, all MutS proteins contain highly conserved ABC-ATPase domains that are essential for MMR ([100] and references therein). Communication between the DNA and the ATP-binding/hydrolysis sites appears to play an important role in the molecular functions of both MutSα and MutSβ [97–99, 101]. In addition to the significant differences between MutSα and MutSβ in substrate recognition and ATP binding and hydrolysis, MutSα differs from MutSβ in the manner that it interacts with PCNA, with MutSα-PCNA complex playing only a limited role in mismatch removal but MutSβ-PCNA complex being highly relevant to small IDLs processing [102]. Finally, current evidence suggests that mammalian MSH2 and MSH6 are transcriptionally and translationally up-regulated in a cell cycle-dependent manner [103, 104], but MSH3 is not cell cycle regulated [104]. (2) DNA repair complex assembly. Subsequent to mismatch binding, MutL proteins are recruited to the MutS proteins/DNA complex in an ATP-dependent manner. Like MutS proteins, MutL proteins function as heterodimers in eukaryotes. The major activity is associated with MutLα (heterocomplex of MLH1-PMS2 in humans but MLH1-PMS1 in yeast and plants). Each subunit of MutLα contains a highly conserved N-terminal region and a weakly conserved C-terminal domain connected by a long flexible linker arm. The N-terminal regions bind and hydrolyze ATP and bind to DNA ([100] and references therein). The C-terminal domains are essential for dimerization [92]. In addition, the C-terminal domains of the PMS subunits have a metal-binding site and an endonuclease activity that is required for MMR [105]. Linker arms vary in length and sequence and are important for MutLα–DNA interactions [ [106] and references therein]. The presence of these linkers has been associated with large asymmetric conformational changes following sequential ATP binding and hydrolysis [107]. (3) Strand discrimination. Identification and discrimination of the newly synthesized DNA strand in eukaryotes are attributed to strand breaks involved in the leading- and lagging-strand synthesis. Replication factor C (RFC) specifically recognizes 3′ termini at the replication fork and loads PCNA asymmetrically onto these sites, with the same side facing towards the strand discontinuity [108]. PCNA loaded at these breaks is required for MutLα endonuclease activation, apparently through protein–protein interaction [109]. Once activated, MutLα incises the strand containing the preexisting strand break due to the loading polarity of PCNA [109]. MutLα endonuclease can also be activated on DNA that contains a MutSβ recognizable lesion but, while MutSα can interact independently with MutLα and PCNA, interactions of MutSβ with MutLα and PCNA were found to be mutually exclusive [102]. These results suggest important differences in the repair mechanisms of single base–base mismatches and IDLs. (4) Strand excision and re-synthesis. Multiply incised molecules are substrates for the 5′–3′ activity of MutSα-activated exonuclease I (Exo1) [110]. Once the mismatch is excised, MutSα and RPA regulate Exo1 activity that leads to excision termination [111]. The repair reaction is completed by a correct DNA re-synthesis by DNA polymerase δ, followed by ligation. In the absence of Exo1, MMR can still be detected suggesting the existence of one or more Exo1-independent modes of mismatch repair. The mechanism most likely involves a strand displacement synthesis by DNA polymerases δ that is strongly stimulated by RPA [112]. Exo1-dependent and independent MMR has been the subject of a recent review [113].
Table 5.
Overview of steps (A) and A. thaliana required factors (B) involved in MMR
MMR in plants
Arabidopsis and other plants encode homologs of MutS and MutL proteins found in other eukaryotic organisms, but also contain a unique MSH polypeptide (MSH7) [114, 115] (Table 5B). MSH7 forms a heterodimer with MSH2 and the protein complex is designated MutSγ. Quantitative analysis of transcript levels of representative genes (MSH2 and PMS1) of the pathway indicated that these genes show a higher expression in calli than in seedlings from A. thaliana, thus confirming the essential genome maintenance function performed by MMR system in rapidly dividing tissues [116, 117]. Further knowledge of the role of a given plant MMR protein came from in vivo studies. Using a yeast system, we have found that AtMutSγ is able to specifically recognize G/T, A/C, T/C, G/A, T/T, or A/A mismatches and/or neighboring sequences containing T [118] and that the expression of AtMutLα affected the repair of IDLs in microsatellite sequences [119]. The functionality of plant MutS and MutL homologs was also assessed using A. thaliana (reviewed in [114]), tomato [120], rice [121] and Nicotiana [122] mutant plants. Suppression of MSH2, PMS1 or MLH1 was associated with microsatellite instability, an increase in homeologous recombination relative to wild-type plants and a broad spectrum of mutant phenotypes in their progenies. Inactivation of MSH7 led a modest increase in the frequency of recombination between Solanum lycopersicum and a distantly related species (Solanum lycopersicoides) [120], while in A. thaliana, the inactivation of MSH7 did not affect the rates of somatic homologous or homeologous recombination, but influenced the rates of meiotic recombination [123]. Finally, somewhat comparable to mammalian proteins, we demonstrated that msh2 mutant plants showed an altered expression pattern of cell cycle marker genes after induced DNA damage in comparison to wild-type plants and we experimentally validated MSH6 as an E2F target gene [124].
Double-strand break repair
Double-strand break (DSB) repair uses two main strategies: a homology-dependent, error-free homologous recombination repair (HR); a potentially error-prone, classical, Ku-dependent non-homologous end-joining (C-NHEJ) and a recently identified error-prone repair pathway, named alternative non-homologous end-joining pathway (A-NHEJ). The choice between these pathways depends on the phase of the cell cycle and the initiation of DNA end resection [125–127].
Homologous recombination
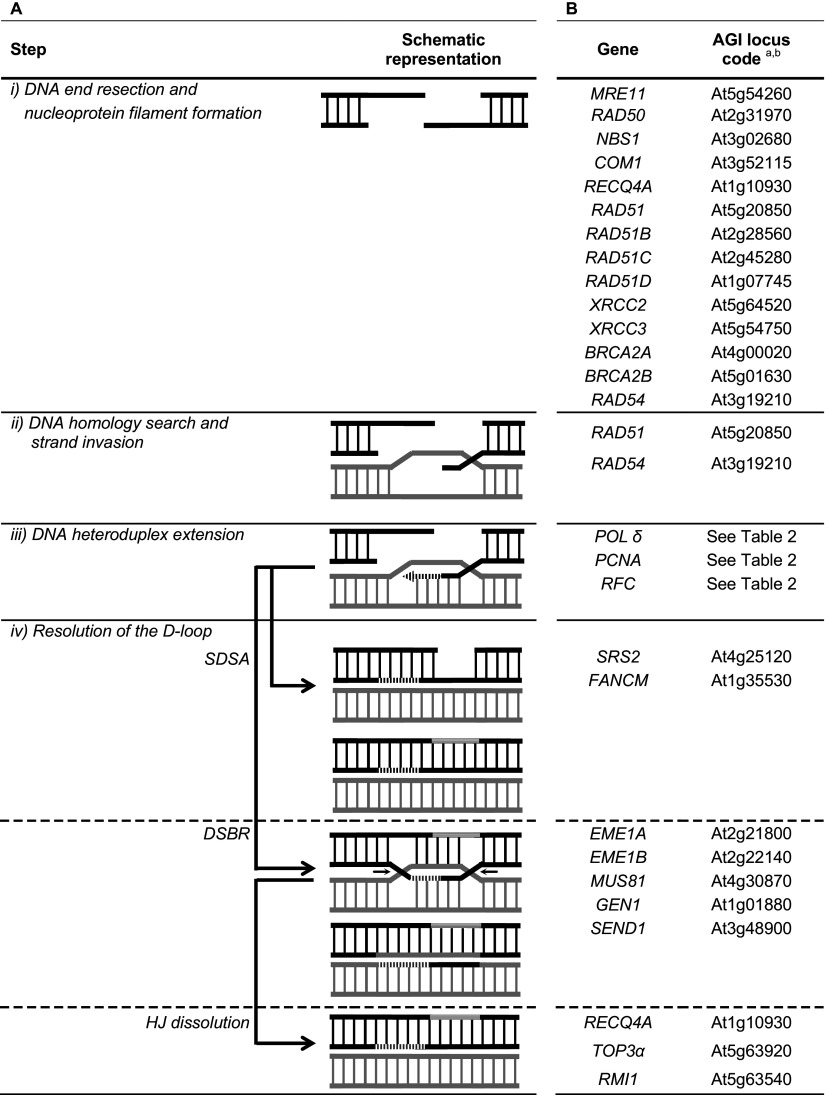
HR is favored during the S/G2 phase of the cell cycle, when sister chromatids are available as homologous templates [128, 129]. This mechanism was the subject of several recent reviews [130–132]. In this pathway, four steps are required (Table 6A). (1) DNA end resection and nucleoprotein filament formation. Initial processing of the ends involves the trimeric complex MRE11-RAD50-NBS1 (MRN) and the CtBP (C-terminal binding protein) interacting protein (CtIP) [133, 134]. Further resection involves the 5′–3′ exonuclease EXO1 or the combined helicase/nuclease activities of the BLM/DNA2 (Bloom Syndrome RecQ Like Helicase/DNA replication helicase/nuclease2) [135]. The 3′ single-strand DNA (ssDNA) tails generated are initially bound by ssDNA-binding protein RPA [136]. Subsequently, RPA is replaced by another ssDNA-binding protein, RAD51, which forms a nucleoprotein filament on the DNA. The replacement of RPA by RAD51 requires several mediator proteins, such as BRCA2 (protein encoded by the breast cancer-associated gene 2) and RAD54 [137–139]. Human cells have two RAD54 homologs, RAD54 and RAD54B, which display similar biochemical activities [140]. Evidence also suggests that RAD51 paralogs, such as XRCC2, XRCC3, RAD51B, RAD51C, and RAD51D, function in promoting and/or stabilizing RAD51 nucleofilaments [131, 141]. However, unlike RAD51 that self-assembles, RAD51 paralogs form at least four different heterodimeric and heterotetrameric complexes. (2) DNA homology search and strand invasion. The RAD51 nucleoprotein filament begins searching for a homologous DNA sequence and then invades the intact double-stranded DNA molecule to form a heteroduplex DNA structure termed the displacement loop (D-loop). D-loop formation is stimulated by RAD54 [131, 132]. Besides ssDNA binding, RAD51 also interacts with dsDNA and associates with the newly formed heteroduplex DNA. (3) DNA heteroduplex extension. The invading strand in the D-loop structure is then extended by several components of the replication machinery, namely DNA polymerase δ, PCNA and RFC [142]. (4) Processing of the D-loop. HR can take several different steps: synthesis-dependent strand annealing (SDSA), Holliday junction (HJ) resolution (the DSBR model) and HJ dissolution [130, 132]. SDSA involves the displacement of the newly synthesized strand by DNA helicase(s), followed by strand annealing, DNA synthesis and ligation. DSBR is characterized by the capture of the second end of the DSB, formation of a double HJ (dHJ) structure and resolution by specialized resolvases (e.g., GEN1, MUS81-EME1, SLX1-SLX4). Alternatively, the dHJ can be dissolved by the BTB complex consisting of BLM helicase, DNA topoisomerase IIIα and BLAP75. Regulation of HJ processing enzymes has been covered in recent reviews [143, 144]. Depending on the pathway employed for HJ processing, different recombination outcomes are generated. SDSA and HJ dissolution lead to the formation of non-crossover while HJ resolution contributes to the formation of non-crossover or crossover. The formation of non-crossover recombinants is promoted in mitotic cells, in contrast with that occurring in meiotic cells.
Table 6.
Overview of steps (A) and A. thaliana required factors (B) involved in HR
a AGI Arabidopsis genome initiative
bAGI locus code for components involved in DNA metabolic pathways is shown in Table 2
An alternative process that involves recombination between regions of homology at both sides of the break can also occur. This process, known as single-strand annealing (SSA), requires HR proteins involved in DNA end resection and annealing and also RPA, but is independent of RAD51 [130–132, 145]. After DNA end resection, the 3′ ssDNA tails generated can reveal complementary sequences on both sides of the break. These sequences anneal forming a branched structure. The 3′ tails are then removed and the nicks are ligated. This process inevitably results in a deletion at the repair junction.
Classical non-homologous end-joining
C-NHEJ functions throughout the cell cycle but is dominant in G1 and G2 [129]. Several recent reviews have focused on C-NHEJ [146–150]. Three different steps are involved in this pathway. (1) DNA end recognition and tethering. The initial step is the recognition and binding of DSBs by the Ku protein (heterodimer of KU70-KU80 in eukaryotes) in a sequence-independent manner. Recent evidence suggests that only two Ku molecules are present at a DSB in vivo, presumably one at each end of the DSB [151]. The Ku-DNA complex then interacts with the DNA-dependent protein kinase (DNA-PK) catalytic subunit (DNA-PKcs) forming a very stable and functional DNA-PK complex. This complex remains tethered to the end and prevents end access. The DNA-PK activated protein undergoes extensive autophosphorylation in vivo [152]. This results in a conformational change that opens the central DNA-binding cavity and releases the autophosphorylated DNA-PKcs from DNA ends [152]. Structures and functions of the Ku protein and the DNA-PK have been recently reviewed [153–155]. (2) DNA end processing. Depending on the nature of the DNA damage, one or more enzyme activities are involved in DNA end processing. For example, the 5′-dRP/AP lyase activity of the Ku protein excises abasic or 5′-dRP/AP sites [156, 157], the tyrosyl DNA phosphodiesterase activity of TDP2 hydrolyzes 5′ phosphotyrosyl-DNA bonds [158], and the 3′-DNA phosphatase and 5′-DNA kinase activities of the polynucleotide kinase/phosphatase (PNKP) generate ligatable 5′-phosphates and 3′-hydroxyl ends [159, 160], among others. In many cases, recruitment of these proteins depends on their direct interaction with Ku [154]. (3) Ligation of the DSB. The final step is carried out by DNA ligase IV, which functions in complex with its cofactors XRCC4 and XLF (XRCC4 like factor, also known as Cernunnos) [150]. Recently, a new protein named PAXX (PAralog of XRCC4 and XLF) has been reported to promote Ku-dependent DNA ligation in vitro [161].
Alternative non-homologous end-joining pathway
A-NHEJ increases as cells enter S-phase [162]. The molecular mechanism of A-NHEJ has recently been reviewed [163–165]. A-NHEJ occurs in the absence of key C-NHEJ factors and can involve alignment of microhomologous sequences; thus, this pathway is also known as microhomology-mediated end-joining (MMEJ). A-NHEJ shares with C-NHEJ similar reaction steps. (1) DNA end recognition and tethering. DSBs are efficiently bound by PARP1 under Ku defective conditions [166]. In fact Ku and PARP1 compete for DNA end binding, and Ku also limits poly ADP-ribose (PAR) synthesis and ssDNA production in response to DSBs [166, 167]. Upon DSB binding, PARP1 gets activated and subsequently auto-poly(ADP-ribosyl)ated. Auto-modification of PARP1 results in a decreased affinity for nucleosomes and in a relaxed local chromatin structure, which facilitates chromatin remodeling for DNA repair [168, 169]. Overviews of the involvement of PARP1 in DNA repair have been published [170–172]. (2) DNA end processing. PARP1-mediated PAR synthesis then triggers recruitment of the MRN complex to initiate incompatible end resection [173, 174]. (3) Ligation of DNA ends. MRN interacts with the DNA ligase IIIα/XRCC1 complex and stimulates the joining of DNA ends [175]. Joining junctions involve microhomologies and deletions [175]. A second pathway independent of microhomologies that relies on DNA ligase I has also been suggested [176].
DSB repair in plants
Knowledge of recombination mechanisms in plants has significantly advanced in recent years, especially because DSB repair emerged as an important tool to achieve controlled modifications of plant genomes (for recent reviews see [177–183]). Most of the HR proteins have been identified and characterized in A. thaliana (Table 6B). Plants with decreased transcript levels of genes involved in the initial processing of the ends, namely MRE11 [22], RAD50 [22], NBS1 [184] and COM1 (a homolog of the human CtIP, [185]) displayed hypersensitivity (measured as a reduction in root growth, fresh weight or development of true leaves) to some genotoxic compounds [mitomycin (MMC) or methylmethanesulfonate (MMS), agents that produce inter-strand DNA crosslinks or that methylates DNA, respectively]. Interaction between NBS1 and MRE11 has been reported for A. thaliana, maize and rice [184, 186]. The NBS1 region involved in the interaction was further characterized in A. thaliana and defined towards the C-terminus [184]. Homologs of BLM, DNA2 and RPA are also conserved in plants. The loss of RECQ4A (a homolog of mammalian BLM helicase, [187–189], JHS1 (a homolog of human DNA2, [190] or RPA1C and RPA1E [89] led to hypersensitivity (measured as a reduction in fresh weight or root growth) in response to a particular DNA damaging agent [MMS; cisplatin (CPT, an agent that mainly induces intra-strand DNA crosslinks); hydroxyurea (an agent that inhibits the ribonucleotide reductase and consequently reduces the dNTP pool and stalls replication forks); zeocin (an agent that induces DSBs) or ionizing radiation]. The recq4A mutant also exhibited an increased frequency of HR [187, 189] suggesting a role for the RECQ4A protein in HR regulation. Suppression of HR is dependent on both the N-terminal region and the helicase activity of the protein [189]. A hyper-recombination phenotype was also observed for the jhs1 mutant, probably due to an increased expression of genes involved in DNA damage repair [190].
Several studies have also addressed the functions of RAD51 and RAD51 paralogues in plants [191–194]. The rad51, rad51c or xrcc3 A. thaliana mutants showed a deficiency in SDSA [192]. Mutants impaired for the other three RAD51 paralogues (RAD51B, RAD51D, XRCC2) were also defective for spontaneous HR [191, 193, 195], although to differing extents [191, 193]. The xrcc2 mutant was the most affected. A further reduction in the recombination rate has been observed in rad51b/xrcc2 double mutant and rad51b/rad51d/xrcc2 triple mutant line [193]. The triple mutant also exhibited a greater sensitivity to the DSB-inducing agent bleomycin (BLE) than the single and double mutants, suggesting a partial functional redundancy [194]. The redundancy of the two AtBRCA2 genes has been also investigated [196–198]. Both Atbrca2a and Atbrca2b mutants showed hypersensitivity against CPT and ionizing irradiation, further stimulated in the Atbrca2a/Atbrca2b double-mutant line [196]. Another report indicated that the double mutant also showed hypersensitivity to MMC and a defect in somatic HR frequency [197].
The role of other factors involved in the process of strand exchange or in the stabilization of recombination intermediates has been elucidated using mutant plants. Loss of RAD54 reduced SDSA and loss of FANCM reduced both SDSA and SSA [192]. Proteins involved in the resolution of the D-loop were characterized in vitro or in vivo. Recombinantly expressed SRS2 protein showed a functional 3′ to 5′ DNA helicase activity that was able to unwind nicked and partial HJ [199]. Biochemical properties of MUS81-EME1 complexes and GEN1 were also determined after cloning, heterologous expression and purification [200, 201]. Two functional EME1 (EME1A and EME1B) and two functional GEN1 [GEN1 (XPG-like endonuclease) and SEND1 (single-strand DNA endonuclease1)] homologs have been identified in A. thaliana. Both complexes, MUS81-EME1A and MUS81-EME1B, were able to cleave nicked HJs and, with a reduced efficiency, intact HJs [201]. Both GEN1 paralogs preferentially cleaved HJs near the junction point; slight differences in sequence preferences have been detected [200]. The role of MUS81 was also defined in vivo. The mus81 mutant line showed a strong reduction in dry weight to MMC, CPT or MMS treatment [188]. Also, altered sensitivity (measured as a reduction in fresh weight) to MMC and CPT was observed in recq4A, top3A and rmi1 single mutant lines [202]. These three lines also showed an enhanced frequency of uninduced somatic HR [202].
In addition to the HR mechanism, KU-dependent and independent pathways have been also identified in plants. Arabidopsis encodes KU70 (At1g16970), KU80 (At1g48050), PARP1 (At2g31320) and PARP2 (At4g02390) [22, 203]. A. thaliana impaired in KU80 showed severe root developmental defects after γ irradiation [204, 205] and higher levels of DSBs [195]. As with the ku80 mutant plant, the xrcc1 mutant line also exhibited a hypersensitivity to γ irradiation [205]. Furthermore, analysis of DSB repair kinetics of ku80 and xrcc1 single mutant lines and ku80/xrcc1 double mutant line indicated a more severe repair defect in the double mutant than in single mutant plants [205]. These observations thus confirm the existence of both KU-dependent and XRCC1-dependent pathways in plants [205]. Besides XRCC1, PARP is also involved in A-NHEJ. In contrast to humans where PARP1 is the major contributor to the PARP activity in response to DNA damage, PARP2 is the primary enzyme responsible for poly(ADP-ribosyl)ation under genotoxic stress in A. thaliana [203]. Plants impaired in both PARP1 and PARP2 displayed a strong reduction in root length and fresh weight after treatment with MMS [206]. However, growth of parp1 and parp2 single mutants was not more sensitive to MMS than wild-type seedlings [206]. Analysis of in vitro end-joining assays indicated that the parp1/parp2 double mutant plant had twofold less end-joining products compared with the wild-type, while the ku70 and ku80 single mutant lines had almost fourfold more products than the wild-type [206]. These observations suggest a regulation of non-homologous recombination pathways. Studies in rice also reported that ku70, ku80, lig4 or xrcc3 mutants showed hypersensitivity to genotoxic agents and/or altered DSB repair [207, 208]. On the other hand, overexpression of OsRecQI4 (the AtRecQ4A counterpart in rice), OsExo1 and MtTpd2a enhanced DSB processing in rice and Medicago truncatula, respectively [209, 210]. No apparent DNA-PK, XLF, DNA ligase III or DNA polymerase β homologs exist in plants [24, 204, 205, 211].
Concluding remarks
Significant progress in plant DNA repair has been made in recent years. Sequence homology-based analysis allowed identification of many plant factors, not only in model organisms but also in some crop species. Most of them were also functionally validated. Thus, it is clear that DNA repair pathways are well conserved between plants and mammals. However, some activities have been particularly intensified in plants, probably related to a better adaptation to variable environmental conditions, from which plants cannot escape, and/or to maintain genome stability over multiple generations. Although only limited information on DNA repair mechanisms from crop plants is available, the current advances in A. thaliana can well be translated to gain insights into the challenges of obtaining new varieties with novel traits.
Acknowledgements
I apologize to those whose work is not mentioned here. I acknowledge research support from Agencia Nacional de Promoción Científica y Tecnológica (PICT 2014-3127). C.P.S. is a member of the Researcher Career of CONICET.
References
- 1.Cadet J, Wagner J. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5:a012559. doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Z, Cools T, De Veylder L. Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol. 2016;67:439–462. doi: 10.1146/annurev-arplant-043015-111902. [DOI] [PubMed] [Google Scholar]
- 4.Yoshiyama K, Sakaguchi K, Kimura S. DNA damage response in plants: conserved and variable response compared to animals. Biology. 2013;2:1338–1356. doi: 10.3390/biology2041338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar B, Zielke N, Gutierrez C. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol. 2014;15:197–210. doi: 10.1038/nrm3756. [DOI] [PubMed] [Google Scholar]
- 6.The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Singh S, Roy S, Choudhury S, Sengupta D. DNA repair and recombination in higher plants: insights from comparative genomics of Arabidopsis and rice. BMC Genom. 2010;11:443. doi: 10.1186/1471-2164-11-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spampinato C, Gómez-Casati D (2012) Research on plants for the understanding of diseases of nuclear and mitochondrial origin. J Biomed Biotech 2012:ID 836196 [DOI] [PMC free article] [PubMed]
- 9.Biedermann S, Mooney S, Hellmann H. Recognition and repair pathways of damaged DNA in higher plants. Selected Topics in DNA Repair. San Diego: University of California; 2011. [Google Scholar]
- 10.Kimura S, Sakaguchi K. DNA repair in plants. Chem Rev. 2006;106:753–766. doi: 10.1021/cr040482n. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg E. A history of the DNA repair and mutagenesis field I. The discovery of enzymatic photoreactivation. DNA Repair (Amst) 2015;33:35–42. doi: 10.1016/j.dnarep.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Eker A, Quayle C, Chaves I, van der Horst G. DNA repair in mammalian cells: direct DNA damage reversal: elegant solutions for nasty problems. Cell Mol Life Sci. 2009;66:968–980. doi: 10.1007/s00018-009-8735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Essen L, Klar T. Light-driven DNA repair by photolyases. Cell Mol Life Sci. 2006;63:1266–1277. doi: 10.1007/s00018-005-5447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancar A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J Biol Chem. 2008;283:32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei Q, Dvornyk V. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS One. 2015;10:e0135940. doi: 10.1371/journal.pone.0135940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okafuji A, Biskup T, Hitomi K, Getzoff E, Kaiser G, Batschauer A, Bacher A, Hidema J, Teranishi M, Yamamoto K, Schleicher E, Weber S. Light-induced activation of class II cyclobutane pyrimidine dimer photolyases. DNA Repair (Amst) 2010;9:495–505. doi: 10.1016/j.dnarep.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Lucas-Lledó J, Lynch M. Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family. Mol Biol Evol. 2009;26:1143–1153. doi: 10.1093/molbev/msp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiontke S, Gnau P, Haselsberger R, Batschauer A, Essen L. Structural and evolutionary aspects of antenna chromophore usage by class II photolyases. J Biol Chem. 2014;289:19659–19669. doi: 10.1074/jbc.M113.542431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rastogi R, Richa X, Kumar A, Tyagi M, Sinha R. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids. 2010;2010:592980. doi: 10.4061/2010/592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richa Sinha R, Häder D. Physiological aspects of UV-excitation of DNA. Top Curr Chem. 2015;356:203–248. doi: 10.1007/128_2014_531. [DOI] [PubMed] [Google Scholar]
- 21.Zhong D. Electron transfer mechanisms of DNA repair by photolyase. Annu Rev Phys Chem. 2015;66:691–715. doi: 10.1146/annurev-physchem-040513-103631. [DOI] [PubMed] [Google Scholar]
- 22.Mannuss A, Trapp O, Puchta H. Gene regulation in response to DNA damage. Biochim Biophys Acta. 2012;1819:154–165. doi: 10.1016/j.bbagrm.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Ueda T, Nakamura C. Ultraviolet-defense mechanisms in higher plants. Biotechnol Biotechnol Equip. 2011;25:2177–2182. doi: 10.5504/BBEQ.2011.0001. [DOI] [Google Scholar]
- 24.Manova V, Gruszka D (2015) DNA damage and repair in plants—from models to crops. Front Plant Sci 6:art 885 [DOI] [PMC free article] [PubMed]
- 25.Li N, Teranishi M, Yamaguchi H, Matsushita T, Watahiki M, Tsuge T, Li S, Hidema J. UV-B-induced CPD photolyase gene expression is regulated by UVR8-dependent and -independent pathways in Arabidopsis . Plant Cell Physiol. 2015;56:2014–2023. doi: 10.1093/pcp/pcv121. [DOI] [PubMed] [Google Scholar]
- 26.Hitomi K, Arvai A, Yamamoto J, Hitomi C, Teranishi M, Hirouchi T, Yamamoto K, Iwai S, Tainer J, Hidema J, Getzoff E. Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J Biol Chem. 2012;287:12060–12069. doi: 10.1074/jbc.M111.244020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hitomi K, DiTacchio L, Arvai A, Yamamoto J, Kim S, Todo T, Tainer J, Iwai S, Panda S, Getzoff E. Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc Natl Acad Sci USA. 2009;106:6962–6967. doi: 10.1073/pnas.0809180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Liu Z, Tan C, Guo X, Wang L, Sancar A, Zhong D. Dynamics and mechanism of repair of UV-induced (6-4) photoproduct by photolyase. Nature. 2010;466:887–890. doi: 10.1038/nature09192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Wang L, Zhong D. Dynamics and mechanisms of DNA repair by photolyase. Phys Chem Chem Phys. 2015;17:11933–11949. doi: 10.1039/C4CP05286B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, Wilson DI. Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krokan H, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svilar D, Goellner E, Almeida K, Sobol R. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks S, Adhikary S, Rubinson E, Eichman B. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs A, Schär P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dianov G, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bebenek K, Pedersen L, Kunkel T. Structure–function studies of DNA polymerase λ. Biochemistry. 2014;53:2781–2792. doi: 10.1021/bi4017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Peña-Diaz J, Andersen S, Slupphaug G, Krokan H, Wilson DI, Akbari M, Otterlei M. XRCC1coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ Mol Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balestrazzi A, Confalonieri M, Macovei A, Donà M, Carbonera D. Genotoxic stress and DNA repair in plants: emerging functions and tools for improving crop productivity. Plant Cell Rep. 2011;30:287–295. doi: 10.1007/s00299-010-0975-9. [DOI] [PubMed] [Google Scholar]
- 39.Roldán-Arjona T, Ariza R. Repair and tolerance of oxidative DNA damage in plants. Mutat Res. 2009;681:169–179. doi: 10.1016/j.mrrev.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Córdoba-Cañero D, Dubois E, Ariza R, Doutriaux M-P, Roldán-Arjona T. Arabidopsis uracil DNA glycosylase (ung) is required for base excision repair of uracil and increases plant sensitivity to 5-fluorouracil. J Biol Chem. 2010;285:7475–7483. doi: 10.1074/jbc.M109.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordoba-Cañero D, Morales-Ruiz T, Roldán-Arjona T, Ariza R. Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 2009;60:716–728. doi: 10.1111/j.1365-313X.2009.03994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Córdoba-Cañero D, Roldán-Arjona T, Ariza R. Arabidopsis ZDP DNA 3′-phosphatase and ARP endonuclease function in 8-oxoG repair initiated by FPG and OGG1 DNA glycosylases. Plant J. 2014;79:824–834. doi: 10.1111/tpj.12588. [DOI] [PubMed] [Google Scholar]
- 43.Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace S, Doublié S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012;11:714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutman B, Niyogi K. Evidence for base excision repair of oxidative DNA damage in chloroplasts of Arabidopsis thaliana . J Biol Chem. 2009;284:17006–17012. doi: 10.1074/jbc.M109.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales-Ruiz T, Ortega-Galisteo A, Ponferrada-Marín M, Martínez-Macías M, Ariza R, Roldán-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponferrada-Marin M, Roldan-Arjona T, Ariza R. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009;37:4264–4274. doi: 10.1093/nar/gkp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brooks S, Fischer R, Huh J, Eichman B. 5-methylcytosine recognition by Arabidopsis thaliana DNA glycosylases DEMETER and DML3. Biochemistry. 2014;53:2525–2532. doi: 10.1021/bi5002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parrilla-Doblas J, Ponferrada-Marin M, Roldan-Arjona T, Ariza R. Early steps of active DNA demethylation initiated by ROS1 glycosylase require three putative helix-invading residues. Nucleic Acids Res. 2013;41:8654–8664. doi: 10.1093/nar/gkt625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong S, Hashimoto H, Kow Y, Zhang X, Cheng X. The carboxy-terminal domain of ROS1 is essential for 5-methylcytosine DNA glycosylase activity. J Mol Biol. 2014;426:3703–3712. doi: 10.1016/j.jmb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramiro-Merina A, Ariza R, Roldán-Arjona T. Molecular characterization of a putative plant homolog of MBD4 DNA glycosylase. DNA Repair (Amst) 2013;12:890–898. doi: 10.1016/j.dnarep.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 51.Chen H, Chu P, Zhou Y, Li Y, Liu J, Ding Y, Tsang E, Jiang L, Wu K, Huang S. Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis . J Exp Bot. 2012;63:4107–4121. doi: 10.1093/jxb/ers093. [DOI] [PubMed] [Google Scholar]
- 52.Nota F, Cambiagno D, Ribone P, Alvarez M. Expression and function of AtMBD4L, the single gene encoding the nuclear DNA glycosylase MBD4L in Arabidopsis . Plant Sci. 2015;235:122–129. doi: 10.1016/j.plantsci.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Qüesta J, Fina J, Casati P. DDM1 and ROS1 have a role in UV-B induced- and oxidative DNA damage in A. thaliana . Front Plant Sci. 2013;4:420. doi: 10.3389/fpls.2013.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy T, Belmonte M, Shu S, Britt A, Hatteroth J. Requirement for abasic endonuclease gene homologues in Arabidopsis seed development. PLoS One. 2009;4:e4297. doi: 10.1371/journal.pone.0004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Córdoba-Cañero D, Qian W, Zhu X, Tang K, Zhang H, Ariza R, Roldán-Arjona T, Zhu J-K. An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis . PLoS Genet. 2015;11:e1004905. doi: 10.1371/journal.pgen.1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cordoba-Cañero D, Roldán-Arjona T, Ariza R. Arabidopsis ARP endonuclease functions in a branched base excision DNA repair pathway completed by LIG1. Plant J. 2011;68:693–702. doi: 10.1111/j.1365-313X.2011.04720.x. [DOI] [PubMed] [Google Scholar]
- 57.Joldybayeva B, Prorok P, Grin I, Zharkov D, Ishenko A, Tudek B, Bissenbaev A, Saparbaev M. Cloning and characterization of a wheat homologue of apurinic/apyrimidinic endonuclease Ape1L. PLoS One. 2014;9:e92963. doi: 10.1371/journal.pone.0092963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martínez-Macías M, Córdoba-Cañero D, Ariza R, Roldán-Arjona T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-mec) excision, gap tailoring, and DNA ligation. J Biol Chem. 2013;288:5496–5505. doi: 10.1074/jbc.M112.427617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martínez-Macías M, Qian W, Miki D, Pontes O, Liu Y, Tang K, Liu R, Morales-Ruiz T, Ariza R, Roldán-Arjona T, Zhu J-K. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis . Mol Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H, Na S, Lee S-Y, Jeong Y-M, Hwang H-J, Hur J, Park S-H, Woo J-C, Kim S. Structure–function studies of a plant tyrosyl-DNA phosphodiesterase provide novel insights into DNA repair mechanisms of Arabidopsis thaliana . Biochem J. 2012;443:49–56. doi: 10.1042/BJ20111308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterworth W, Kozak J, Provost C, Bray C, Angelis K, West C. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009;9:79. doi: 10.1186/1471-2229-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Wen C, Liu S, Zheng L, Shen B, Tao Y. Shade avoidance 6 encodes an Arabidopsis flap endonuclease required for maintenance of genome integrity and development. Nucleic Acids Res. 2016;44:1271–1284. doi: 10.1093/nar/gkv1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alekseev S, Coin F. Orchestral maneuvers at the damaged sites in nucleotide excision repair. Cell Mol Life Sci. 2015;72:2177–2186. doi: 10.1007/s00018-015-1859-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schärer O. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spivak G, Ganesan A. The complex choreography of transcription-coupled repair. DNA Repair (Amst) 2014;19:64–70. doi: 10.1016/j.dnarep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 66.Vermeulen W, Fousteri M. Mammalian transcription coupled excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bedez F, Linard B, Brochet X, Ripp R, Thompson J, Moras D, Lecompte O, Poch O. Functional insights into the core-TFIIH from a comparative survey. Genomics. 2013;101:178–186. doi: 10.1016/j.ygeno.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Fuss J, Tainer J. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 2011;10:697–713. doi: 10.1016/j.dnarep.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fagbemi A, Orelli B, Schärer O. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2011;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogi T, Limsirichaikul S, Overmeer R, Volker M, Takenaka K, Cloney R, Nakazawa Y, Nimi A, Jaspers N, Mullenders L, S Y, Fousteri M, Lehamann A (2010) Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 37:714–727 [DOI] [PubMed]
- 71.Ganpudi A, Schroeder D, editors. UV damaged DNA repair and tolerance in plants. Intech, Croatia: Selected Topics in DNA Repair; 2011. [Google Scholar]
- 72.Farmer L, Book A, Lee K, Lin Y, Fu H, Vierstra R. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis . Plant Cell. 2010;22:124–142. doi: 10.1105/tpc.109.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al Khateeb W, Schroeder D. DDB2, DDB1A and DET1 exhibit complex interactions during Arabidopsis development. Genetics. 2007;176:231–242. doi: 10.1534/genetics.107.070359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernhardt A, Mooney S, Hellmann H. Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta. 2010;232:555–566. doi: 10.1007/s00425-010-1195-9. [DOI] [PubMed] [Google Scholar]
- 75.Zhang C, Guo H, Zhang J, Guo G, Schumaker K, Guo Y. Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell. 2010;22:2353–2369. doi: 10.1105/tpc.110.073973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biedermann S, Hellmann H. The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana . Plant J. 2010;62:404–415. doi: 10.1111/j.1365-313X.2010.04157.x. [DOI] [PubMed] [Google Scholar]
- 77.Molinier J, Lechner E, Dumbliauskas E, Genschik P. Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 2008;4:e1000093. doi: 10.1371/journal.pgen.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Al Khateeb W, Schroeder D. Overexpression of Arabidopsis damaged DNA binding protein 1A (DDB1A) enhances UV tolerance. Plant Mol Biol. 2009;70:371–383. doi: 10.1007/s11103-009-9479-9. [DOI] [PubMed] [Google Scholar]
- 79.Ganpudi A, Schroeder D (2013) Genetic interactions of Arabidopsis thaliana damaged DNA binding protein 1B (DDB1B) with DDB1A, DET1, and COP1. G3 (Bethesda) 3:493–503 [DOI] [PMC free article] [PubMed]
- 80.Koga A, Ishibashi T, Kimura S, Uchiyama Y, Sakaguchi K. Characterization of T-DNA insertion mutants and RNAi silenced plants of Arabidopsis thaliana UV-damaged DNA binding protein 2 (AtUV-DDB2) Plant Mol Biol. 2006;61:227–240. doi: 10.1007/s11103-006-6408-z. [DOI] [PubMed] [Google Scholar]
- 81.Ly V, Hatherell A, Kim E, Chan A, Belmonte M, Schroeder D. Interactions between Arabidopsis DNA repair genes UVH6, DDB1A, and DDB2 during abiotic stress tolerance and floral development. Plant Sci. 2013;213:88–97. doi: 10.1016/j.plantsci.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 82.Shaked H, Avivi-Ragolsky N, Levy A. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang L, Flury S, Kalck V, Hohn B, Molinier J. CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol Biol. 2006;61:345–356. doi: 10.1007/s11103-006-0016-9. [DOI] [PubMed] [Google Scholar]
- 84.Vonarx E, Tabone E, Osmond M, Anderson H, Kunz B. Arabidopsis homologue of human transcription factor IIH/nucleotide excision repair factor p44 can function intranscription and DNA repair and interacts with AtXPD. Plant J. 2006;46:512–521. doi: 10.1111/j.1365-313X.2006.02705.x. [DOI] [PubMed] [Google Scholar]
- 85.Gentile A, Ditt R, Dias F, Da Silva M, Dornelas M, Menossi M. Characterization of ScMat1, a putative TFIIH subunit from sugarcane. Plant Cell Rep. 2009;28:663–672. doi: 10.1007/s00299-008-0663-1. [DOI] [PubMed] [Google Scholar]
- 86.Gutierrez C. The Arabidopsis cell division cycle. Arabidopsis Book. 2009;7:e0120. doi: 10.1199/tab.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, De Bodt S, Maere S, Laukens K, Pharazyn A, Ferreira P, Eloy N, Renne C, Meyer C, Faure J, Steinbrenner J, Beynon J, Larkin J, Van de Peer Y, Hilson P, Kuiper M, De Veylder L, Van Onckelen H, Inzé D, Witters E, De Jaeger G. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana . Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eschbach V, Kobbe D. Different replication protein A complexes of Arabidopsis thaliana have different DNA-binding properties as a function of heterotrimer composition. Plant Cell Physiol. 2014;55:1460–1472. doi: 10.1093/pcp/pcu076. [DOI] [PubMed] [Google Scholar]
- 89.Aklilu B, Soderquist R, Culligan K. Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 2014;42:3104–3108. doi: 10.1093/nar/gkt1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aklilu B, Culligan K. Molecular evolution and functional diversification of Replication Protein A1 in plants. Front Plant Sci. 2016;7:33. doi: 10.3389/fpls.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peña-Diaz J, Jiricny J. Mammalian mismatch repair: error-free or error-prone? Trends Biochem Sci. 2012;37:206–214. doi: 10.1016/j.tibs.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 92.Kunkel T, Erie D. Eukaryotic mismatch repair in relation to DNA replication. Annu Rev Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reyes G, Schmidt T, Kolodner R, Hombauer H. New insights into the mechanism of DNA mismatch repair. Chromosoma. 2015;124:443–462. doi: 10.1007/s00412-015-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Friedhoff P, Li P, Gotthardt J. Protein-protein interactions in DNA mismatch repair. DNA Repair (Amst) 2016;38:50–57. doi: 10.1016/j.dnarep.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 95.Kolodner R. A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair (Amst) 2016;38:3–13. doi: 10.1016/j.dnarep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sachadyn P. Conservation and diversity of MutS proteins. Mutat Res. 2010;694:20–30. doi: 10.1016/j.mrfmmm.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 97.Tian L, Gu L, Li G-M. Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSa and MutSb determine their differential mismatch binding activities. J Biol Chem. 2009;284:11557–11562. doi: 10.1074/jbc.M900908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Warren J, Pohlhaus T, Changela A, Iyer R, Modrich P, Beese L. Structure of the human MutSα DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 99.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat Struct Mol Biol. 2011;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Groothuizen F, Sixma T. The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst) 2016;38:14–23. doi: 10.1016/j.dnarep.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 101.Owen B, Lang W, McMurray C. The nucleotide binding dynamics of human MSH2-MSH3 are lesion dependent. Nat Struct Mol Biol. 2009;16:550–557. doi: 10.1038/nsmb.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iyer R, Pluciennik A, Genschel J, Tsai M, Beese L, Modrich P. MutLαand proliferating cell nuclear antigen share binding sites on MutSβ. J Biol Chem. 2010;285:11730–11739. doi: 10.1074/jbc.M110.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Edelbrock M, Kaliyaperumal S, Williams K. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat Res. 2013;743–744:53–66. doi: 10.1016/j.mrfmmm.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mjelle R, Hegre S, Aas P, Slupphaug G, Drabløs F, Saetrom P, Krokan H. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst) 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 105.Kadyrova L, Kadyrov F. Endonuclease activities of MutLα and its homologs in DNA mismatch repair. DNA Repair (Amst) 2016;38:42–49. doi: 10.1016/j.dnarep.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guarné A, Charbonnier J. Insights from a decade of biophysical studies on MutL: roles in strand discrimination and mismatch removal. Prog Biophys Mol Biol. 2016;117:149–156. doi: 10.1016/j.pbiomolbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 107.Sacho E, Kadyrov F, Modrich P, Kunkel T, Erie D. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutLα. Mol Cell. 2008;29:112–121. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McNally R, Bowman G, Goedken E, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pluciennik A, Dzantiev L, Iyer R, Constantin N, Kadyrov F, Modrich P. PCNA function in the activation and strand direction of MutLαendonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shao H, Baitinger C, Soderblom E, Burdett V, Modrich P. Hydrolytic function of Exo1 in mammalian mismatch repair. Nucleic Acids Res. 2014;42:7104–7112. doi: 10.1093/nar/gku420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Genschel J, Modrich P. Functions of MutLalpha, replication protein A (RPA), and HMGB1 in 5′-directed mismatch repair. J Biol Chem. 2009;284:21536–21544. doi: 10.1074/jbc.M109.021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kadyrov F, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci USA. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goellner E, Putnam C, Kolodner R. Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst) 2015;32:24–32. doi: 10.1016/j.dnarep.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spampinato C, Gomez R, Galles C, Lario L. From bacteria to plants: a compendium of mismatch repair assays. Mutat Res. 2009;682:110–128. doi: 10.1016/j.mrrev.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 115.Tam S, Samipak S, Britt A, Chetelat R. Characterization and comparative sequence analysis of the DNA mismatch repair MSH2 and MSH7 genes from tomato. Genetica. 2009;137:341–354. doi: 10.1007/s10709-009-9398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Galles C, Gomez R, Spampinato C. PMS1 from Arabidopsis thaliana: optimization of protein overexpression in Escherichia coli . Mol Biol Rep. 2011;38:1063–1070. doi: 10.1007/s11033-010-0203-2. [DOI] [PubMed] [Google Scholar]
- 117.Gomez R, Galles C, Spampinato C. High-level production of MSH2 from Arabidopsis thaliana: a DNA mismatch repair system key subunit. Mol Biotechnol. 2011;47:120–129. doi: 10.1007/s12033-010-9319-9. [DOI] [PubMed] [Google Scholar]
- 118.Gomez R, Spampinato C. Mismatch recognition function of Arabidopsis thaliana MutSγ. DNA Repair (Amst) 2013;12:257–264. doi: 10.1016/j.dnarep.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 119.Galles C, Spampinato C. Yeast mutator phenotype enforced by Arabidopsis PMS1 expression. Mol Biol Rep. 2013;40:2107–2114. doi: 10.1007/s11033-012-2269-5. [DOI] [PubMed] [Google Scholar]
- 120.Tam S, Hays J, Chetelat R. Effects of suppressing the DNA mismatch repair system on homeologous recombination in tomato. Theor Appl Genet. 2011;123:1445–1458. doi: 10.1007/s00122-011-1679-4. [DOI] [PubMed] [Google Scholar]
- 121.Xu J, Li M, Chen L, Wu G, Li H. Rapid generation of rice mutants via the dominant negative suppression of the mismatch repair protein OsPMS1. Theor Appl Genet. 2012;125:975–986. doi: 10.1007/s00122-012-1888-5. [DOI] [PubMed] [Google Scholar]
- 122.Van Marcke I, Angenon G. Genomic stability in Nicotiana plants upon silencing of the mismatch repair gene MSH2 . Plant Biotechnol Rep. 2013;7:467–480. doi: 10.1007/s11816-013-0285-0. [DOI] [Google Scholar]
- 123.Lario L, Botta P, Casati P, Spampinato C. Role of AtMSH7 in UV-B-induced DNA damage recognition and recombination. J Exp Bot. 2015;66:3019–3026. doi: 10.1093/jxb/eru464. [DOI] [PubMed] [Google Scholar]
- 124.Lario L, Ramirez-Parra E, Gutierrez C, Casati P, Spampinato C. Regulation of plant MSH2 and MSH6 genes in the UV-B-induced DNA damage response. J Exp Bot. 2011;62:2925–2937. doi: 10.1093/jxb/err001. [DOI] [PubMed] [Google Scholar]
- 125.Ceccaldi R, Rondinelli B, D’Andrea A. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez B. Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res. 2012;2:249–268. [PMC free article] [PubMed] [Google Scholar]
- 127.Kakarougkas A, Jeggo P. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young J, Tkác J, Cook M, Rosebrock A, Munro M, Canny M, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 129.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kowalczykowski S. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2015;7:a016410. doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sartori A, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson S. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Takeda S, Nakamura K, Taniguchi Y, Paull T. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell. 2007;28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 135.Nimonkar A, Genschel J, Kinoshita E, Polaczek P, Campbell J, Wyman C, Modrich P, Kowalczykowski S. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huertas P, Jackson S. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;264:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carreira A, Kowalczykowski S. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci USA. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reuter M, Zelensky A, Smal I, Meijering E, van Cappellen W, de Gruiter H, van Belle G, van Royen M, Houtsmuller A, Essers J, Kanaar R, Wyman C. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J Cell Biol. 2014;207:599–613. doi: 10.1083/jcb.201405014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Agarwal S, van Cappellen W, Guénolé A, Eppink B, Linsen S, Meijering E, Houtsmuller A, Kanaar R, Essers J. ATP-dependent and independent functions of Rad54 in genome maintenance. J Cell Biol. 2011;192:735–750. doi: 10.1083/jcb.201011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ceballos S, Heyer W. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta. 2011;1809:509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol. 2011;22:898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 142.Sneeden J, Grossi S, Tappin I, Hurwitz J, Heyer W. Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic Acids Res. 2013;41:4913–4925. doi: 10.1093/nar/gkt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Blanco M, Matos J. Hold your horSSEs: controlling structure-selective endonucleases MUS81 and Yen1/GEN1. Front Genet. 2015;6:253. doi: 10.3389/fgene.2015.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matos J, West S. Holliday junction resolution: regulation in space and time. DNA Repair (Amst) 2014;19:176–181. doi: 10.1016/j.dnarep.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez B. Is homologous recombination really an error-free process? Front Genet. 2014;5:175. doi: 10.3389/fgene.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bétermier M, Bertrand P, Lopez B. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chiruvella K, Liang Z, Wilson T. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Radhakrishnan S, Jette N, Lees-Miller S. Non-homologous end joining: emerging themes and unanswered questions. DNA Repair (Amst) 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Waters C, Strande N, Wyatt D, Pryor J, Ramsden D. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst) 2014;17:39–51. doi: 10.1016/j.dnarep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Williams G, Hammel M, Radhakrishnan S, Ramsden D, Lees-Miller S, Tainer J. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Britton S, Coates J, Jackson S. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol. 2013;202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dobbs T, Tainer J, Lees-Miller S. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Davis A, Chen B, Chen D. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fell V, Schild-Poulter C. The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res. 2015;763:15–29. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 155.Jette N, Lees-Miller S. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Roberts S, Strande N, Burkhalter M, Strom C, Havener J, Hasty P, Ramsden D. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Strande N, Roberts S, Oh S, Hendrickson E, Ramsden D. Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends. J Biol Chem. 2012;287:13686–13693. doi: 10.1074/jbc.M111.329730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pommier Y, Huang S, Gao R, Das B, Murai J, Marchand C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair (Amst) 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Coquelle N, Havali-Shahriari Z, Bernstein N, Green R, Glover J. Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single and double-stranded DNA substrates. Proc Natl Acad Sci USA. 2011;108:21022–21027. doi: 10.1073/pnas.1112036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Garces F, Pearl L, Oliver A. The structural basis for substrate recognition by mammalian polynucleotide kinase 3′ phosphatase. Mol Cell. 2011;44:385–396. doi: 10.1016/j.molcel.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ochi T, Blackford A, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, Wu Q, Draviam V, Robinson C, Blundell T, Jackson S. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Truong L, Li Y, Shi L, Hwang P, He J, Wang H, Razavian N, Berns M, Wu X. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci USA. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Decottignies A. Alternative end-joining mechanisms: a historical perspective. Front Genet. 2013;4:48. doi: 10.3389/fgene.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Frit P, Barboule N, Yuan Y, Gomez D, Calsou P. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst) 2014;17:81–97. doi: 10.1016/j.dnarep.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 165.Sfeir A, Symington L. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cheng Q, Barboule N, Frit P, Gomez D, Bombarde O, Couderc B, Ren G, Salles B, Calsou P. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res. 2011;39:9605–9619. doi: 10.1093/nar/gkr656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Muthurajan U, Hepler M, Hieb A, Clark N, Kramer M, Yao T, Luger K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci USA. 2014;111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Polo S, Jackson S. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329:18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 171.Ko H, Ren E. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules. 2012;2:524–548. doi: 10.3390/biom2040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pines A, Mullenders L, van Attikum H, Luijsterburg M. Touching base with PARPs: moonlighting in the repair of UV lesions and double-strand breaks. Trends Biochem Sci. 2013;38:321–330. doi: 10.1016/j.tibs.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 173.Bryant H, Petermann E, Schultz N, Jemth A, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Haince J, McDonald D, Rodrigue A, Dery U, Masson J, Hendzel M, Poirier G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 175.Della-Maria J, Zhou Y, Tsai M, Kuhnlein J, Carney J, Paull T, Tomkinson A. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Simsek D, Brunet E, Wong S-W, Katyal S, Gao Y, McKinnon P, Lou J, Zhang L, Li J, Rebar E, Gregory P, Holmes M, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Baltes N, Voytas D. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 2015;33:120–131. doi: 10.1016/j.tibtech.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 178.Knoll A, Fauser F, Puchta H. DNA recombination in somatic plant cells: mechanisms and evolutionary consequences. Chromosome Res. 2014;22:191–201. doi: 10.1007/s10577-014-9415-y. [DOI] [PubMed] [Google Scholar]
- 179.Gaj T, Gersbach C, Cr Barbas. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Voytas D. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol. 2013;64:327–350. doi: 10.1146/annurev-arplant-042811-105552. [DOI] [PubMed] [Google Scholar]
- 181.Kumar S, Barone P, Smith M. Gene targeting and transgene stacking using intra genomic homologous recombination in plants. Plant Methods. 2016;12:11. doi: 10.1186/s13007-016-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Puchta H, Fauser F. Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 2014;78:727–741. doi: 10.1111/tpj.12338. [DOI] [PubMed] [Google Scholar]
- 183.Sprink T, Metje J, Hartung F. Plant genome editing by novel tools: tALEN and other sequence specific nucleases. Curr Opin Biotechnol. 2015;32:47–53. doi: 10.1016/j.copbio.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 184.Waterworth W, Altun C, Armstrong S, Roberts N, Dean P, Young K, Weil C, Bray C, West C. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007;52:41–52. doi: 10.1111/j.1365-313X.2007.03220.x. [DOI] [PubMed] [Google Scholar]
- 185.Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlögelhofer P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Akutsu N, Iijima K, Hinata T, Tauchi H. Characterization of the plant homolog of Nijmegen breakage syndrome 1: involvement in DNA repair and recombination. Biochem Biophys Res Commun. 2007;353:394–398. doi: 10.1016/j.bbrc.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 187.Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana . Proc Natl Acad Sci USA. 2007;104:18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mannuss A, Dukowic-Schulze S, Suer S, Hartung F, Pacher M, Puchta H. RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana . Plant Cell. 2010;22:3318–3330. doi: 10.1105/tpc.110.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schröpfer S, Kobbe D, Hartung F, Knoll A, Puchta H. Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 2014;42:1684–1697. doi: 10.1093/nar/gkt1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jia N, Liu X, Gao H. A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol. 2016;171:318–333. doi: 10.1104/pp.16.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Da Ines O, Degroote F, Amiard S, Goubely C, Gallego M, White C. Effects of XRCC2 and RAD51B mutations on somatic and meiotic recombination in Arabidopsis thaliana . Plant J. 2013;74:959–970. doi: 10.1111/tpj.12182. [DOI] [PubMed] [Google Scholar]
- 192.Roth N, Klimesch J, Dukowic-Schulze S, Pacher M, Mannuss A, Puchta H. The requirement for recombination factors differs considerably between different pathways of homologous double-strand break repair in somatic plant cells. Plant J. 2012;72:781–790. doi: 10.1111/j.1365-313X.2012.05119.x. [DOI] [PubMed] [Google Scholar]
- 193.Serra H, Da Ines O, Degroote F, Gallego M, White C. Roles of XRCC2, RAD51B and RAD51D in RAD51-independent SSA recombination. PLoS Genet. 2013;9:e1003971. doi: 10.1371/journal.pgen.1003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang Y, Xiao R, Wang H, Cheng Z, Li W, Zhu G, Wang Y, Ma H. The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation. New Phytol. 2014;201:292–304. doi: 10.1111/nph.12498. [DOI] [PubMed] [Google Scholar]
- 195.Yao Y, Bilichak A, Titov V, Golubov A, Kovalchuk I. Genome stability of Arabidopsis atm, ku80 and rad51b mutants: somatic and transgenerational responses to stress. Plant Cell Physiol. 2013;54:982–989. doi: 10.1093/pcp/pct051. [DOI] [PubMed] [Google Scholar]
- 196.Abe K, Osakabe K, Ishikawa Y, Tagiri A, Yamanouchi H, Takyuu T, Yoshioka T, Ito T, Kobayashi M, Shinozaki K, Ichikawa H, Toki S. Inefficient double-strand DNA break repair is associated with increased fasciation in Arabidopsis BRCA2 mutants. J Exp Bot. 2009;60:2751–2761. doi: 10.1093/jxb/erp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Seeliger K, Dukowic-Schulze S, Wurz-Wildersinn R, Pacher M, Puchta H. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana . New Phytol. 2012;193:364–375. doi: 10.1111/j.1469-8137.2011.03947.x. [DOI] [PubMed] [Google Scholar]
- 198.Trapp O, Seeliger K, Puchta H. Homologs of breast cancer genes in plants. Front Plant Sci. 2011;2:19. doi: 10.3389/fpls.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Blanck S, Kobbe D, Hartung F, Fengler K, Focke M, Puchta H. A SRS2 homolog from Arabidopsis thaliana disrupts recombinogenic DNA intermediates and facilitates single strand annealing. Nucleic Acids Res. 2009;37:7163–7176. doi: 10.1093/nar/gkp753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bauknecht M, Kobbe D. AtGEN1 and AtSEND1, two paralogs in Arabidopsis, possess holliday junction resolvase activity. Plant Physiol. 2014;166:202–216. doi: 10.1104/pp.114.237834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Geuting V, Kobbe D, Hartung F, Dürr J, Focke M, Puchta H. Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol. 2009;150:1062–1071. doi: 10.1104/pp.109.136846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana . PLoS Genet. 2008;4:e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Song J, Keppler B, Wise R, Bent A. PARP2 is the predominant Poly(ADP-Ribose) polymerase in Arabidopsis DNA damage and immune responses. PLoS Genet. 2015;11:e1005200. doi: 10.1371/journal.pgen.1005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Charbonnel C, Allain E, Gallego M, White C. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis . DNA Repair (Amst) 2011;10:611–619. doi: 10.1016/j.dnarep.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 205.Charbonnel C, Gallego M, White C. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010;64:280–290. doi: 10.1111/j.1365-313X.2010.04331.x. [DOI] [PubMed] [Google Scholar]
- 206.Jia Q, den Dulk-Ras A, Shen H, Hooykaas P, de Pater S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana . Plant Mol Biol. 2013;82:339–351. doi: 10.1007/s11103-013-0065-9. [DOI] [PubMed] [Google Scholar]
- 207.Nishizawa-Yokoi A, Nonaka S, Saika H, Kwon Y, Osakabe K, Toki S. Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol. 2012;196:1048–1059. doi: 10.1111/j.1469-8137.2012.04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang B, Wang M, Tang D, Li Y, Xu M, Gu M, Cheng Z, Yu H. XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis. J Exp Bot. 2015;66:5713–5725. doi: 10.1093/jxb/erv253. [DOI] [PubMed] [Google Scholar]
- 209.Confalonieri M, Faè M, Balestrazzi A, Donà M, Macovei A, Valassi A, Giraffa G, Carbonera D. Enhanced osmotic stress tolerance in Medicago truncatula plants overexpressing the DNA repair gene MtTdp2a (tyrosyl-DNA phosphodiesterase 2) Plant Cell Tiss Organ Cult. 2014;116:187–203. doi: 10.1007/s11240-013-0395-y. [DOI] [PubMed] [Google Scholar]
- 210.Kwon Y, Abe K, Osakabe K, Endo M, Nishizawa-Yokoi A, Saika H, Shimada H, Toki S. Overexpression of OsRecQl4 and/or OsExo1 enhances DSB-induced homologous recombination in rice. Plant Cell Physiol. 2012;53:2142–2152. doi: 10.1093/pcp/pcs155. [DOI] [PubMed] [Google Scholar]
- 211.Yoshiyama K. Recent progress in research on DNA damage responsesin animals and plants. Genes Genet Syst. 2016;90:185–186. doi: 10.1266/ggs.15-10001. [DOI] [PubMed] [Google Scholar]