Abstract
The α-amylase is a ubiquitous starch hydrolase catalyzing the cleavage of the α-1,4-glucosidic bonds in an endo-fashion. Various α-amylases originating from different taxonomic sources may differ from each other significantly in their exact substrate preference and product profile. Moreover, it also seems to be clear that at least two different amino acid sequences utilizing two different catalytic machineries have evolved to execute the same α-amylolytic specificity. The two have been classified in the Cabohydrate-Active enZyme database, the CAZy, in the glycoside hydrolase (GH) families GH13 and GH57. While the former and the larger α-amylase family GH13 evidently forms the clan GH-H with the families GH70 and GH77, the latter and the smaller α-amylase family GH57 has only been predicted to maybe define a future clan with the family GH119. Sequences and several tens of enzyme specificities found throughout all three kingdoms in many taxa provide an interesting material for evolutionarily oriented studies that have demonstrated remarkable observations. This review emphasizes just the three of them: (1) a close relatedness between the plant and archaeal α-amylases from the family GH13; (2) a common ancestry in the family GH13 of animal heavy chains of heteromeric amino acid transporter rBAT and 4F2 with the microbial α-glucosidases; and (3) the unique sequence features in the primary structures of amylomaltases from the genus Borrelia from the family GH77. Although the three examples cannot represent an exhaustive list of exceptional topics worth to be interested in, they may demonstrate the importance these enzymes possess in the overall scientific context.
Keywords: α-Amylase family GH13, Plant and archaeal α-amylases, Heavy-chains of rBAT and 4F2 proteins, Family GH77 amylomaltases of borrelian origin, Evolutionary relatedness
Introduction
α-Amylase (EC 3.2.1.1) represents probably the best known and most deeply studied amylolytic enzyme [1–6]. It catalyzes the hydrolytic cleavage of the α-1,4-glucosidic linkages in starch and related α-glucans in an endo-fashion employing the retaining reaction mechanism. Its evolution started to attract the serious scientific interest approximately 25 years ago when it became clear that there is a group of starch hydrolases and related enzymes possessing closely related functions within the frame of homologous amino acid sequences [7–9]. It was, for example, the enzyme cyclodextrin glucanotransferase (CGTase; EC 2.4.1.19) sharing with the α-amylase the first step of the catalyzed reaction, which in the CGTase proceeds not with the molecule of water like in the α-amylase but with a molecule of a saccharide to be transferred [10]. For CGTases, it was thus revealed that, despite obvious differences between an α-amylase and a CGTase, they exhibit interesting sequence similarities with α-amylases [11] that previously might even complicate the correct assignment of the α-amylase/CGTase specificity [12–14] as well as the answer concerning their evolutionary history [15]. The other closely related enzyme was the so-called maltohexaohydrolase (EC 3.2.1.98), i.e., the exo-amylase producing maltohexaose [16]. The discovery of a completely novel enzyme, the neopullulanase (EC 3.2.1.135) in 1989 [17], able to perform both hydrolysis and transglycosylation of both the α-1,4- and α-1,6-glucosidic linkages [18–20], can be considered as a milestone in creating the overall view of the group of sequentially and functionally related amylolytic enzymes. Thus, also from the evolutionary point of view, a new enzyme family, named according to its leading member as the α-amylase family, has been established [20–24].
Interestingly, from the very beginning the α-amylase family has offered various surprises or at least the phenomena deserving a special attention. For example, even the α-amylases originating from various sources (roughly from Bacteria, Archaea and Eucarya) may differ from each other quite substantially in their substrate preference and/or product profile although they are still active towards and produce various α-glucans [1]. Some of them have been confirmed to be able to transglycosylate to a small, limited extent, e.g., the α-amylase from Pseudoalteromonas haloplanktis [25].
Historically, the establishment of the four conserved sequence regions (CSRs) among the primary structures of α-amylases in 1986 [26] has played an unique role, especially as one of the requirements for an enzyme to become a member of the family [20]. Soon after the four CSRs were observed in other enzymes (members) of the developing α-amylase family [8, 9, 20, 22] and completed by the three additional CSRs [27–29]. Currently, it has been recommended to use as many as possible of the seven defined CSRs [30, 31] to best characterize a protein as a member of the α-amylase family, mainly if there are any doubts concerning the exact enzyme specificity [1, 32].
Almost simultaneously with the gradual appearance and final definition of the α-amylase enzyme family in the literature early in 1990s [7–9, 20–24], a pioneering study was published [33] delivering the newly developed concept of classification of glycoside hydrolases (GHs) to sequence-based families, placing the group of enzymes known already at that time as the α-amylase family into the family GH13. After a few published updates [34, 35], the increasing system of GH families has been incorporated into the current web-server database CAZy (Carbohydrate-Active enzymes) [36], the α-amylase enzyme specificity being potentially found also in families GH57, GH119 and even GH126 [1, 37]. Nevertheless, the family GH13 represents the main α-amylase family [1] and with approximately 31,500 sequences and more than 30 different enzyme specificities (Table 1) it belongs—among 129 GH functional families created until now—to the largest GH families at all [37]. On a higher level of hierarchy, it forms the clan GH-H together with families GH70 and GH77 [31, 37, 38]; the former of these two families covers various glucansucrases with typically circularly permuted primary structure with respect to that seen in the main family GH13 [39–41], whereas the latter is the monospecific family of 4-α-glucanotransferases known also as amylomaltase and disproportionating enzyme (DPE) in prokaryotes and eukaryotes, respectively [42–44]. On the lower level of hierarchy, in 2006 the family GH13, reflecting also the previous efforts [45], was officially divided into 35 subfamilies by the CAZy curators [46]; currently the number of GH13 subfamilies has reached 41 [37, 47, 48] and it will very probably raise in the future [49].
Table 1.
Members of the α-amylase GH families
| Enzyme class | Enzyme | EC no. | GH family | GH13 subfamily |
|---|---|---|---|---|
| Hydrolases | α-Amylase | 3.2.1.1 | 13, 57, 119, 126 | 1, 5, 6, 7, 15, 20, 24, 27, 28, 32, 36, 37 |
| Oligo-1,6-glucosidase | 3.2.1.10 | 13 | 23, 31 | |
| α-Glucosidase | 3.2.1.20 | 13 | 17, 21, 23, 30, 31, 40 | |
| α-Galactosidase | 3.2.1.22 | 57 | ||
| Pullulanase | 3.2.1.41 | 13 | 12, 13, 14 | |
| Amylopullulanase | 3.2.1.1/41 | 13, 57 | 12, 14, 39 | |
| Cyclomaltodextrinase | 3.2.1.54 | 13, 57 | 20, 36 | |
| Maltotetraose-forming amylase | 3.2.1.60 | 13 | 19 | |
| Isoamylase | 3.2.1.68 | 13 | 11, 14 | |
| Isoamylase/4-α-glucanotransferase | 3.2.1.68/2.4.1.25 | 13 | 11 | |
| Dextran glucosidase | 3.2.1.70 | 13 | 31 | |
| Trehalose 6-phosphate hydrolase | 3.2.1.93 | 13 | 29 | |
| Maltohexaose-forming amylase | 3.2.1.98 | 13 | 5, 19 | |
| Maltotriose-forming amylase | 3.2.1.116 | 13 | 2, 32 | |
| Maltogenic amylase | 3.2.1.133 | 13, 57 | 2, 20 | |
| Neopullulanase | 3.2.1.135 | 13 | 20 | |
| Maltooligosyltrehalose threhalohydrolase | 3.2.1.141 | 13 | 10 | |
| Sucrose hydrolase | 3.2.1.– | 13 | 4 | |
| Maltopentaose-forming amylase | 3.2.1.– | 13 | 5 | |
| Glycogen degrading enzyme | 3.2.1.– | 13 | 12 | |
| Cyclic α-maltosyl-1,6-maltose hydrolase | 3.2.1.– | 13 | 20 | |
| Transferases | Amylosucrase | 2.4.1.4 | 13 | 4 |
| Dextransucrase | 2.4.1.5 | 70 | ||
| Sucrose phosphorylase | 2.4.1.7 | 13 | 18 | |
| Glucan branching enzyme | 2.4.1.18 | 13, 57 | 8, 9 | |
| Cyclodextrin glucanotransferase | 2.4.1.19 | 13 | 2 | |
| 4-α-Glucanotransferase | 2.4.1.25 | 13, 57, 77 | ||
| Glucan debranching enzyme | 2.4.1.25/3.2.1.33 | 13 | 11, 20, 25 | |
| Alternansucrase | 2.4.1.140 | 70 | ||
| α-1,3-Glucan synthase | 2.4.1.183 | 13 | 22 | |
| α-1,4-Glucan: phosphate α-maltosyltransferase | 2.4.99.16 | 13 | 3 | |
| Sucrose-6-phosphate phosphorylase | 2.4.1.– | 13 | 18 | |
| α-Transglucosidase | 2.4.1.– | 13 | 23 | |
| Isocyclomaltooligosaccharide glucanotransferase | 2.4.1.– | 13 | ||
| α-4,6-Glucanotransferase | 2.4.1.– | 70 | ||
| Reuteran sucrase | 2.4.1.– | 70 | ||
| α-1,6/α-1,2-Branching glucansucrase | 2.4.1.– | 70 | ||
| Isomerases | Isomaltulose synthase | 5.4.99.11 | 13 | 31 |
| Maltooligosyltrehalose synthase | 5.4.99.15 | 13 | 26 | |
| Trehalose synthase | 5.4.99.16 | 13 | 16, 33 | |
| HAT proteins | hc-rBAT protein | – | 13 | 35 |
| 4F2hc antigen | – | 13 | 34 |
As far as the family GH13 is concerned in its entirety, it has become evident that the same enzyme specificity (e.g., the α-amylase or pullulanase) may exist in several separated groups or GH13 subfamilies, but on the other hand some other specificities (e.g., oligo-1,6-glucosidase, α-glucosidase and dextran glucosidase) may exist altogether within a single group or GH13 subfamily. The attractiveness of the α-amylase clan GH-H for scientists has been strengthened also by classifying into the family the heavy-chains of heteromeric amino acid transporters, known as rBAT protein and 4F2 antigen (i.e., neither amylolytic enzymes, nor any enzymes at all) that, being typically of animal origin, are evolutionary related to bacterial α-glucosidases [50]. The present review aims to deliver the updated view of a few perhaps most remarkable evolutionary relatedness observed within the α-amylase family.
The family GH13 α-amylases from plants and archaea
While the first amino acid sequences of the α-amylases from Archaea have become available only at the end of 1990s [51–55], the primary structures of their first plant counterparts were determined at least 10–15 years earlier [56–60]. Moreover, the three-dimensional structure of the high pI isozyme of the barley α-amylase AMY-2 was solved a few years before [61] the first archaeal α-amylase sequence from Pyrococcus furiosus was announced [51, 52]. The barley α-amylase AMY-2 structure [61], solved later also as a complex with acarbose [62], was at that time only the fourth known α-amylase tertiary structure [30], in addition to those from Aspergillus oryzae [63, 64], Aspergillus niger [65, 66] and pig pancreas [67, 68]. At present, with regard to plant and archaeal α-amylases, tertiary structures have been solved and published also for the low pI isozyme of the barley α-amylase AMY-1 [69, 70], the α-amylase from rice [71, 72] as well as the α-amylase from Pyrococcus woesei [73].
From the evolutionary point of view, before the α-amylases from archaea were known, plant α-amylases formed a compact cluster neighboring with the liquefying α-amylases from bacilli [28]. As documented by various evolutionarily oriented studies [73–78] this has remained true until now with a significant upgrading of the original picture [28] illustrating that the cluster of archaeal α-amylases shares the branch with that of plant counterparts [53, 79]. According to the CAZy nomenclature [46], the α-amylases from plants and archaea have been assigned the subfamily number GH13_6 and GH13_7, respectively; the bacterial homologues representing mostly liquefying α-amylases from bacilli being classified within the subfamily GH13_5. It should be pointed out that all these three subfamilies of α-amylases are mutually very closely related and the exact branching pattern among them in the evolutionary tree may depend on the aligned segment of their amino acid sequences (i.e., just CSRs, the catalytic domain or the entire sequence, etc.) [76].
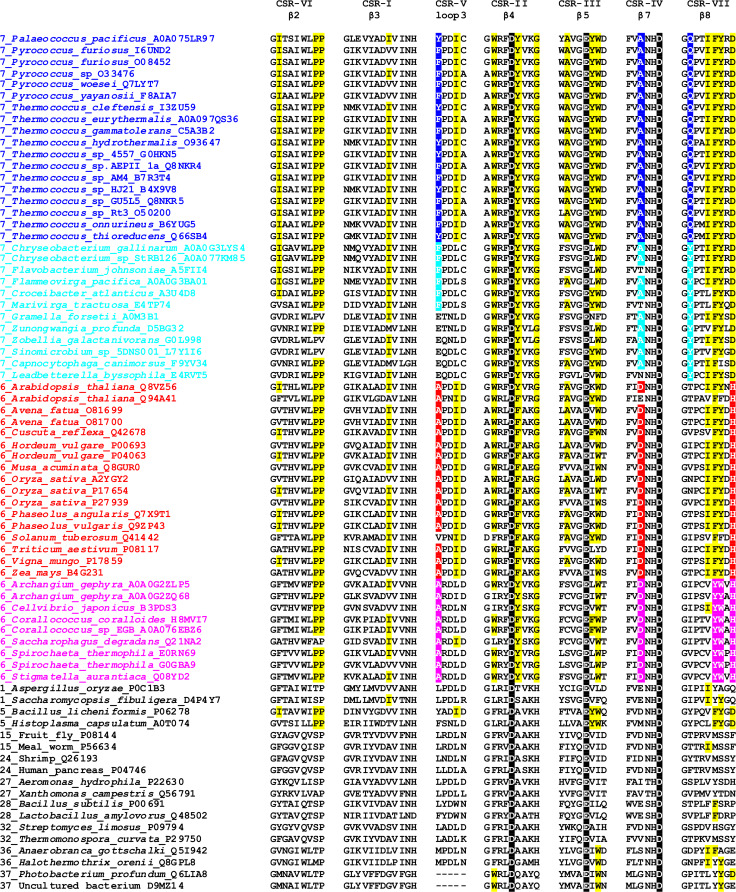
The pronounced relatedness between the plant and archaeal α-amylases is remarkable not only due to their long taxonomical distance (eukaryotic plants and prokaryotic archaea) but also when differences in their thermostability (or the temperature optimal for their enzymatic activity) are considered [1]. The close positions in the evolutionary tree obviously reflect the similarities throughout their amino acid sequences, especially within the CSRs (Fig. 1) described first in detail in 1999 [79]. While the sequence similarities shared by both plant and archaeal α-amylases represent something that distinguishes both subfamilies from remaining GH13 α-amylases, there have to be some additional sequence features that are unique for each group, i.e., that discriminate the two subfamilies from each other, just these unique sequence features should be used in the efforts aimed at identifying the factors that could be responsible for the hyperthermostability of archaeal α-amylases (or, on the other hand, the low thermostability of the α-amylases from plants). Thus, for example, the two residues deserving the attention in the α-amylase from Thermococcus hydrothermalis could be as follows (Fig. 1): (1) the first position of the CSR-V, i.e., Tyr184Ala (184_YPDIC in the T. hydrothermalis α-amylase versus 146_APDID in the barley high pI α-amylase isozyme), because among the archaea it is occupied only by either phenylalanine or tyrosine (i.e., an aromatic residue); and (2) the third position of the CSR-IV, i.e., Ala308Asp (306_FVANHD versus 284_FVDNHD), because there is invariantly conserved alanine among the archaea in comparison with invariant aspartic acid in plants.
Fig. 1.
Comparison of seven CSRs of the family GH13 α-amylases with focus on the subfamilies GH13_6 and GH13_7. These CSRs cover mostly individual β-strands of the catalytic TIM-barrel domain [32]. Each α-amylase in the list is characterized by the GH13 subfamily number [46], the source of origin (the organism) and the accession number from the UniProt database [221]. The α-amylases from the subfamilies GH13_6 and GH13_7 were collected from the actual CAZy database [37], whereas the set of representative α-amylases from the GH13 remaining subfamilies was prepared according to previous in silico studies [74–79]. The catalytic triad is signified by black-and-white inversion. Sequence features characteristic of the GH13_7 archaeal α-amylases are highlighted in yellow. The features typical specifically for each group of α-amylases within the two GH13 subfamilies, i.e., (1) archaea and (flavo)-bacteria within GH13_7; and (2) plants and bacteria within GH13_6, which may discriminate the individual groups from each other are emphasized by respective colors
Recently, some hypothetical bacterial (i.e., not archaeal) α-amylases from genome sequencing projects were assigned to the “archaeal” subfamily GH13_7 [1, 37]. Currently, in the CAZy database, all these belong to the phylum Flavobacteria [37]. It is, however, worth mentioning that even a simple BLAST search [80] using the flavobacterial GH13_7 α-amylase from Sinomicrobium sp. 5DNS001 [81] as a query retrieves hypothetical bacterial α-amylases with GH13_7 sequence features, but not belonging to Flavobacteria. Anyhow, the moderately thermostable Sinomicrobium sp. 5DNS001 α-amylase [81], clearly homologous to its archaeal hyperthermostable counterparts (Fig. 2), may add to our understanding of the rules that have governed the evolution of plant, (flavo)-bacterial and archaeal α-amylases and the factors that are responsible for their thermostability differences [76, 79, 81–83]. In analogy with the above-mentioned features in which the plant and archaeal α-amylases differ from each other, it makes sense to try to identify the features in the amino acid sequences of (flavo)-bacterial α-amylases that are well conserved but simultaneously well discriminating them from their archaeal counterparts (Fig. 1). One example is the second position of the CSR-VII, i.e., Tyr363Gln (362_GYPTVFYGD in the Sinomicrobium sp. 5DNS001 α-amylase versus 330_GQPAIFYRD in the T. hydrothermalis α-amylase isozyme), because both the tyrosine and the glutamine are invariantly conserved among the respective groups of bacterial and archaeal α-amylases. Thus, a silico analysis as detailed as possible focused on comparison of amino acid sequences is of special importance, especially if, at the tertiary structure level the individual representatives of both GH13_6 and GH13_7 subfamilies look very similar and obviously without any substantial differences (Fig. 2).
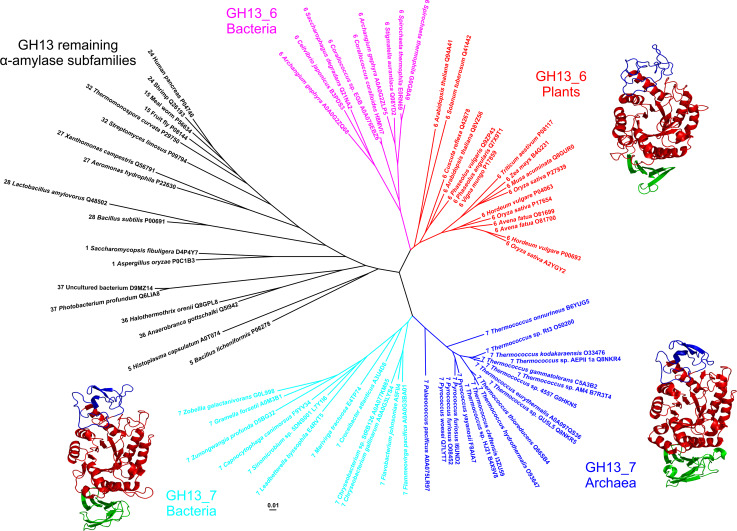
Fig. 2.
Evolutionary tree of the family GH13 α-amylases with focus on the subfamilies GH13_6 and GH13_7. The tree is based on the alignment of seven conserved sequence regions, shown in Fig. 1. The tree was calculated using the neighbor-joining clustering [222] implemented in the Clustal-W2 phylogeny package [223] available at the European Bioinformatics Institute’s web-site (http://www.ebi.ac.uk/), and then displayed with the program iTOL [224]. Tertiary structures of representatives of flavobacterial, archeal and plant family GH13 α-amylases are shown near their clusters in the tree. Sources of the α-amylases: Sinomicrobium sp. 5DNS001 [81] (subfamily GH13_7; flavobacteria); Pyrococcus woesei [72] (subfamily GH13_7; archaea); Hordeum vulgare—barley high pI isozyme AMY-2 [61] (subfamily GH13_6; plants). The archaeal and plant α-amylases are experimentally solved crystal structures retrieved from the Protein Data Bank (PDB) [225] under the PDB codes 1MWO and 1AMY, respectively. The flavobacterial α-amylase is a tertiary structure model obtained at the fold recognition server Phyre-2 [89] for its amino acid sequence (UniProt accession number: L7Y1I6; residues: Gly52-Gly477) based on the P. woesei α-amylase structure (1MWO) as template. The individual domains are colored as follows: catalytic (β/α)8-barrel—red, domain B—blue, domain C—green. The structural models were displayed with the program PyMol [226]
It is worth mentioning that, in addition to archaeal α-amylases from the class Thermococci (namely the two genera Pyrococcus and Thermococcus) classified within the subfamily GH13_7, there are several potential α-amylases produced by halophilic archaea (the class Halobacteria) deserving the attention that, however, until now have not been assigned any GH13 subfamily [37]. Some of them simply may not be true α-amylases, e.g., those from Natronococcus amylolyticus [84, 85] and Haloarcula japonica [86] producing mainly maltotriose and maltose, respectively. For others, the ambiguity arises from the fact that the studied sequences originate from genome projects [87], i.e., they are still hypothetical amylolytic enzymes. Interestingly, for the sequence of the halophilic α-amylase from Haloarcula hispanica [88] the fold recognition server Phyre-2 [89] revealed the Bacillus stearothermophilus α-amylase from the subfamily GH13_5 [90] as the best structural template.
Animal transport proteins rBAT and 4F2 versus the microbial GH13 α-glucosidases
A sequence resemblance of heavy-chains of the heteromeric amino acid transporters (hcHATs), the rBAT protein and the 4F2 antigen, to α-glucosidases from the α-amylase family was originally revealed already in 1992 in two independent studies [91, 92]. However, for a few subsequent years it was, in fact, not seriously taken into account by researchers engaged in the enzymology of the α-amylase family. The main reason was that the hcHAT proteins obviously do not obey all the criteria defined for an enzyme to be a member of the α-amylase family [93, 94]. The eventual evolutionary relatedness between the hcHATs and the α-amylase family enzymes was first studied in a detail in 1997 [95] when the attention was paid mainly to domain B, protruding typically in the family GH13 out of the catalytic (β/α)8-barrel (simply called the TIM-barrel [1, 30, 31]) between the strand β3 and the helix α3 [61, 63, 67, 72, 96–104], which in the rBAT protein is present, whereas the 4F2 antigen lacks it (Fig. 3a). Both the rBAT protein and 4F2 antigen belong to the so-called solute carrier families SLC7 (the light chain) and SLC3 (the heavy chain); the respective hetero-chains being connected by a disulphide bridge [105–109]. Both groups have been accepted as members of the CAZy α-amylase family GH13 members and received the subfamily numbers GH13_34 and GH13_35 for the heavy chains of 4F2 antigens and rBAT proteins, respectively [46]. HAT proteins are responsible for transport of various types of amino acids across the plasma membrane in animals, namely in mammals, and their defects may lead to a failure in amino acids re-absorption and digestion (e.g., cystinuria, lysinuric protein intolerance) [107–109]. While the light chain has nothing to do with the family GH13 (it is a hydrophobic transmembrane protein consisting of 12 α-helices), the heavy subunit consists of the intracellular N-terminus, a transmembrane region and a large extracellular C-terminal part that exhibits the unambiguous sequence-structural similarity with α-glucosidases from the α-amylase family GH13 [91, 92, 110, 111]. Thus, the transportation activity is operated by the light subunit, whereas the heavy chain functions as a chaperone to help to orient the light chain to a proper position in the plasma membrane [112–114]. Note that, only the heavy subunit in its C-terminal part exhibits clear sequence similarities with the members of the α-amylase family [50, 91, 92, 95, 103, 104].
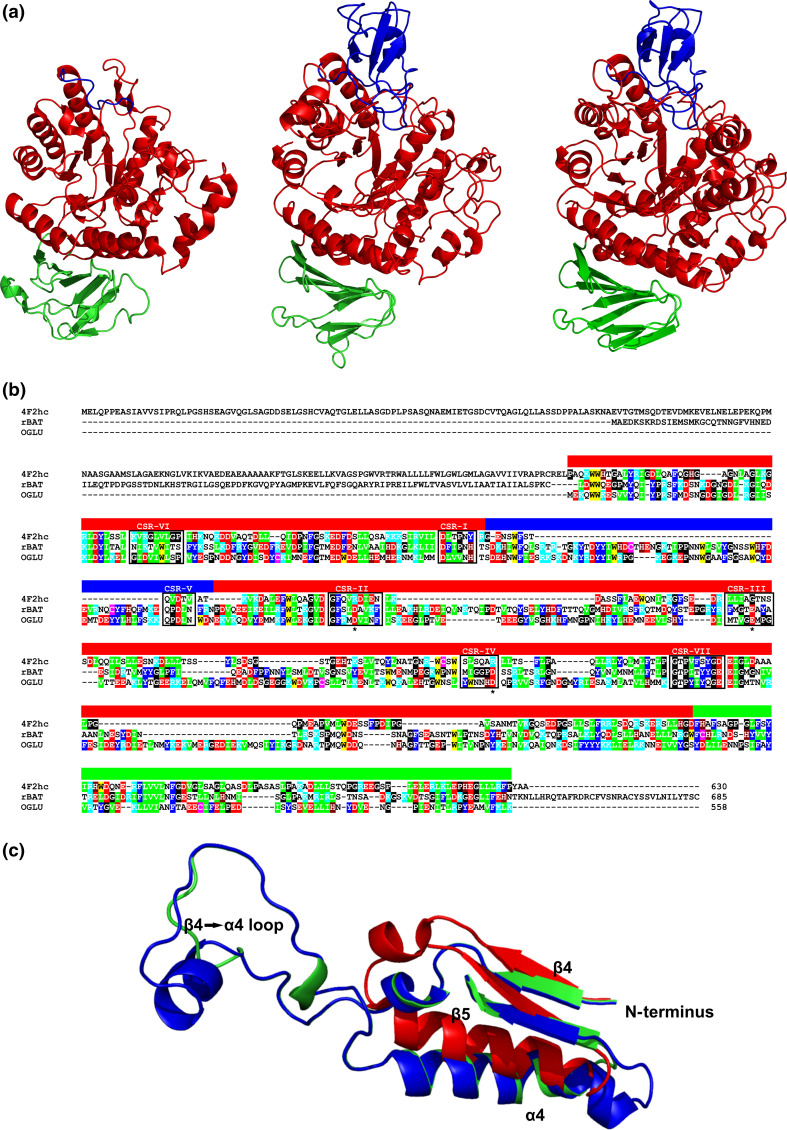
Fig. 3.
a Comparison of tertiary structures of the family GH13 hcHAT proteins from animals and α-glucosidase from bacteria. Sources of the proteins: Homo sapiens 4F2hc antigen [103] (subfamily GH13_34; left); Homo sapiens rBAT protein [111] (subfamily GH13_35; middle); Bacillus cereus oligo-1,6-glucosidase [96] (subfamily GH13_31; right). b Amino acid sequence alignment of the same three proteins: human 4F2hc antigen [103] (UniProt accession No.: P08195-1); human rBAT protein [111] (Q07837); Bacillus cereus oligo-1,6-glucosidase (OGLU) [96] (P21332). Color code for the selected residues: W, yellow; F, Y—blue; V, L, I—green; D, E—red; R, K—cyan; H—brown; C—magenta; G, P—black. The seven characteristic CSRs are boxed and marked above the alignment. The colored lane above the alignment blocks means the three domains shown in a. The catalytic triad is signified by asterisks under the alignment. c Structural overlay emphasizing the longer loop 4 connecting the strand β4 to the helix α4 in the oligo-1,6-glucosidase (green) and rBAT protein (blue) in comparison to a very short version present in the 4F2hc antigen (red). The structures were superimposed using the MultiProt web-server [227] (http://bioinfo3d.cs.tau.ac.il/MultiProt/); the overlap being characterized by 280 corresponding Cα-atoms and the RMSD value of 0.96 Å. The human 4F2hc antigen and bacterial oligo-1,6-glucosidase are experimentally solved crystal structures retrieved from the PDB [225] under the PDB codes 2DH3 and 1UOK, respectively. The human rBAT protein is a tertiary structure model obtained at the fold recognition server Phyre-2 [89] for its amino acid sequence (UniProt accession number: Q07837; residues: Asp116-Glu649) based on the B. cereus oligo-1,6-glucosidase structure (1UOK) as template. The individual domains are colored as follows: catalytic (β/α)8-barrel—red, domain B—blue, domain C—green. The structures were visualized with the program PyMol [226]
Interestingly, however, the heavy chains of both the rBAT protein and the 4F2 antigen display the best similarity to the α-amylase family GH13 enzymes in the parts of their sequences that are not involved in the catalytic action (Fig. 3b), i.e., in CSR-VI (the strand β2 of the catalytic TIM-barrel) and CSR-VII (the strand β8) [50]. While the heavy chain of the 4F2 antigen does not possess the segment corresponding with the domain B [50, 95, 103], the heavy subunit of the rBAT protein contains the entire domain B that is, moreover, very closely related to its counterpart seen in the members of the so-called oligo-1,6-glucosidases [45, 48, 95], sharing even the sequence fingerprint QPDLN of the CRS-V [50, 95]. The evidently closer relationships between the enzymes from the oligo-1,6-glucosidase subfamily and the heavy-chains of the rBAT proteins is reflected also in the presence of catalytic residues in the transport proteins, although in all cases the entire GH13 catalytic triad is not conserved completely. This feature could even be traced among the potential sequences of the heavy subunits of the rBAT proteins originated from basal metazoa [50]. For the heavy chain of the 4F2 antigen, no α-glucosidase activity has been detected [103], which is in agreement with a lack of residues that would correspond to the catalytic triad of the members of the α-amylase family [50, 103, 104], i.e., the aspartic acid as the catalytic nucleophile at the strand β4 (the CSR-II), the glutamic acid as the proton donor at the strand β5 (the CSR-III) and the aspartic acid as the transition-state stabilizer at the strand β7 (the CSR-IV) [1, 31].
With regard to sequence-structural resemblance of heavy chains of both the rBAT protein and 4F2 antigen, it is worth mentioning that, for example, the human 4F2 heavy chain lacks not only domain B, but also a stretch of about 40 amino acid residues succeeding the strand β4, i.e., it possesses a very short loop 4 connecting the strand β4 to helix α4 [50], whereas in both the oligo-1,6-glucosidase and the human rBAT heavy chain (both having also the entire domain B [93, 102]) the loop connecting the strand β4 to the helix α4 is longer (Fig. 3c). It is of note that even the GH13 neopullulanase subfamily members [45, 115], possessing shorter domain B [95], also lack the longer excursion of the loop 4 segment, but currently it is still unknown whether or not the domain B in GH13 oligo-1,6-glucosidase subfamily (and also in rBATs) operates in conjunction with the prolonged loop 4 (the β4 to α4 connection) [50]. It seems, however, that the consecutive loss of domain B in the heavy subunits of the 4F2 antigens might be connected with adequate shortening of the loop4, since this observation can be generalized to all 4F2 heavy-chains proteins [50]. Noticeably, similar heteromeric amino acid transporter system known in mammals was observed in insect [116, 117], nematodes [118] and schistozomes [119]. According to the in silico analysis of hcHATs and their enzymatic counterparts from the α-amylase family GH13 [50], a protein that could be close to the common ancestor of hcHATs and GH13 α-glucosidases might be represented by the hypothetical GH13-like protein from the cnidarian starlet sea anemone Nematostella vectensis.
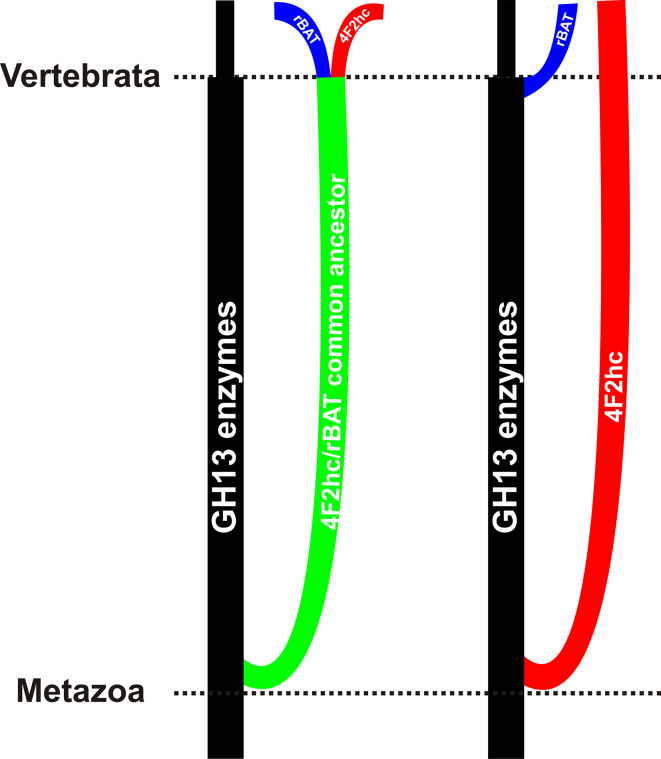
Due to the obviously closer similarity of the heavy chains of rBAT proteins to the family GH13 α-glucosidases than that of those of 4F2 antigens, it is not easy to draw an unambiguous scenario of their evolution [95]. Based on a detailed in silico analysis [50], two different scenarios should be considered (Fig. 4). One of the possibilities means that the division of hcHATs from the enzymes of the α-amylase family might occur in one single event in basal Metazoa and subsequent split to rBAT proteins and 4F2 antigens in chordates. The above-mentioned higher similarity of the rBAT proteins to the GH13 enzymes than to the 4F2hc antigens could be explained by a selection pressure acting against change upon rBAT (and not upon 4F2hc), to preserve its eventual enzymatic capabilities, although until now no evidence has been delivered concerning the enzymatic activity of any rBAT protein [50]. Alternatively, the 4F2hc protein could be experiencing accelerated evolution toward some new function, accompanied by changes in both sequence and structure, while rBAT has retained its original function, remained mostly unchanged and thus similar to GH13 enzymes. The other eventuality assumes two independent branching events: one of the 4F2 antigens in the basal Metazoa and the other one of the rBAT proteins directly from enzymes of oligo-1,6-glucosidase subfamily of the α-amylase family in chordates. Since chordates obviously do not possess enzymes of the oligo-1,6-glucosidase subfamily [45, 48, 50], their α-glucosidases counterparts could have been transformed into the heavy chains of the rBAT proteins [120]. Both scenarios (Fig. 4) reflect the ancestry of both rBAT proteins and 4F2 antigens anchored within the GH13 α-amylase family, the difference being only in the way leading from the GH13 enzymes either to rBAT and 4F2 together or to rBAT and 4F2 separately [50]. At present, it seems that the hcHATs are found in animals starting from basal Metazoa, but it is not possible to exclude that in the future some new sequences of hcHAT-like proteins of non-metazoan origin become available [121].
Fig. 4.
Evolutionary scenarios of hcHAT proteins with respect to the α-amylase family GH13. The left eventuality considers a single-event division of both rBAT proteins and 4F2hc antigens from the enzymes of the α-amylase family in basal Metazoa with a subsequent split to rBAT proteins and 4F2 antigens in higher animals like chordates. The right possibility counts with two independent evolutionary events leading first to separation of the 4F2hc antigens from the α-amylase family enzymes in the basal Metazoa and second to recruitment of the rBAT proteins from enzymatic members of the oligo-1,6-glucosidase subfamily in higher animals
The family GH77 amylomaltases from borreliae
The family GH77, together with families GH13 and GH70, is a member of the α-amylase clan GH-H [31, 37]. As already mentioned above, it is, in contrast to the main α-amylase family GH13, a monospecific family, i.e., it contains only one enzyme specificity of 4-α-glucanotransferase (EC 2.4.1.25) [122]. The trivial name of the 4-α-glucanotransferase within the family GH77 has been distinguished with regard to taxonomy—while in prokaryotes (both Bacteria and Archaea) the name amylomaltase has been used, the name disproportionating enzyme (DPE) has been established in eukaryotes (mainly in plants and green algae) [42–44, 123–135]. In general, the 4-α-glucanotransferase catalyzes, employing the retaining reaction mechanism, the intermolecular transglycosylation of α-1,4-glucans, i.e., it transfers a glucan chain from one α-glucan to another one or within a single linear glucan molecule to produce a cyclic α-1,4-glucan [42–44, 136–138]. Currently, the family counts almost 3000 sequenced members with absolute domination of Bacteria accompanied approximately equally by a few dozens each from Archaea and Eucarya [37].
From the structural point of view, since the family GH77 is a member of the α-amylase clan GH-H, the GH77 4-α-glucanotransferase possesses a typical catalytic (β/α)8-barrel (TIM-barrel). It contains just more insertions in comparison with the family GH13 TIM-barrel having, in fact, only domain B protruding out of the barrel in the place of the loop 3 connecting the strand β3 to the helix α3 (Fig. 2). The family GH77 4-α-glucanotransferase TIM-barrel has thus three subdomains called B1, B2 and B3 (Fig. 5), of which B1 and B3 correspond with domains B and C in the family GH13, while the subdomain B2 is unique for the family GH77 [44, 139–143]. It is, however, worth mentioning that although the subdomain B3 may play the role of the C-terminal family GH13 domain C succeeding the catalytic TIM-barrel, it is not an antiparallel β-sandwich (a Greek key motif) seen typically in the GH13 (see Figs. 2, 5).
Fig. 5.
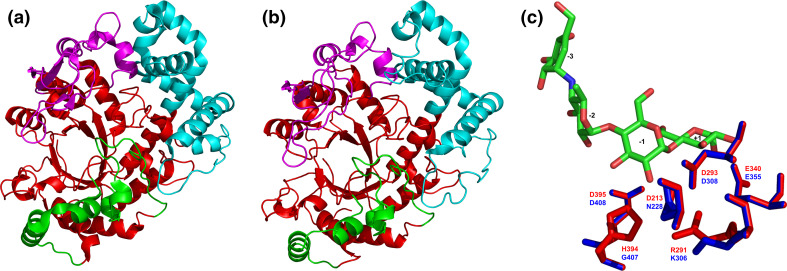
Tertiary structures of the family GH77 amylomaltases from bacteria and their comparison. Sources of the amylomaltases: a Thermus aquaticus [139]; b Borrelia burgdorferi [129]. c Structural overlay focusing on the active-site residues with complexed acarbose in the Thermus amylomaltase (red) compared with the naturally mutated corresponding residues in the Borrelia amylomaltase (blue). The structures were superimposed using the MultiProt web-server [227] (http://bioinfo3d.cs.tau.ac.il/MultiProt/); the overlap being characterized by 488 corresponding Cα-atoms and the RMSD value of 0.18 Å. The Thermus amylomaltase is experimentally solved crystal structure retrieved from the PDB [225] under the PDB code 1ESW, whereas the Borrelia amylomaltase is a tertiary structure model obtained at the homology modeling server SwissModel [228] for its amino acid sequence (UniProt accession number: A6YM39; residues: Asn12-Ala507) based on the T. aquaticus amylomaltase structure (1CWY [44]) as template. The individual domains are colored as follows: catalytic (β/α)8-barrel—red, subdomain B1—cyan, subdomain B2—magenta, subdomain B3—green. The exact positions of displayed active-site residues in the amino acid sequence can be identified in the alignment shown in Fig. 6. Based on the accepted nomenclature [229] the acarbose occupies the subsites from −3 to +1. The structures were visualized with the program PyMol [226]
Within the family GH77, the amylomaltases from the genus Borrelia obviously play a special (evolutionary) role [122]. Originally, based on the complete genome sequence of the Lyme disease spirochaete Borrelia burgdorferi [144], an in silico analysis published in 2003 [145] delivered a remarkable observation of mutations in several conserved and functionally important positions in the B. burgdorferi hypothetical amylomaltase encoded by the gene malQ. The importance of the eventual mutations in the hypothetical GH77 amylomaltase was strengthened by the fact that, one of the residues was the otherwise throughout the entire clan GH-H invariantly conserved arginine in the position i-2 with respect to catalytic nucleophile (aspartic acid) located at the strand β4 of the catalytic TIM-barrel, i.e., in the CSR-II [145]. The possible implications of remarkable in silico observations thus evoked a serious interest for experimental confirmation. The gene malQ from B. burgdorferi was cloned and expressed in Escherichia coli and the recombinant amylomaltase was shown to exhibit not only all the unique sequence features seen in the hypothetical MalQ but also the amylomaltase activity [129].
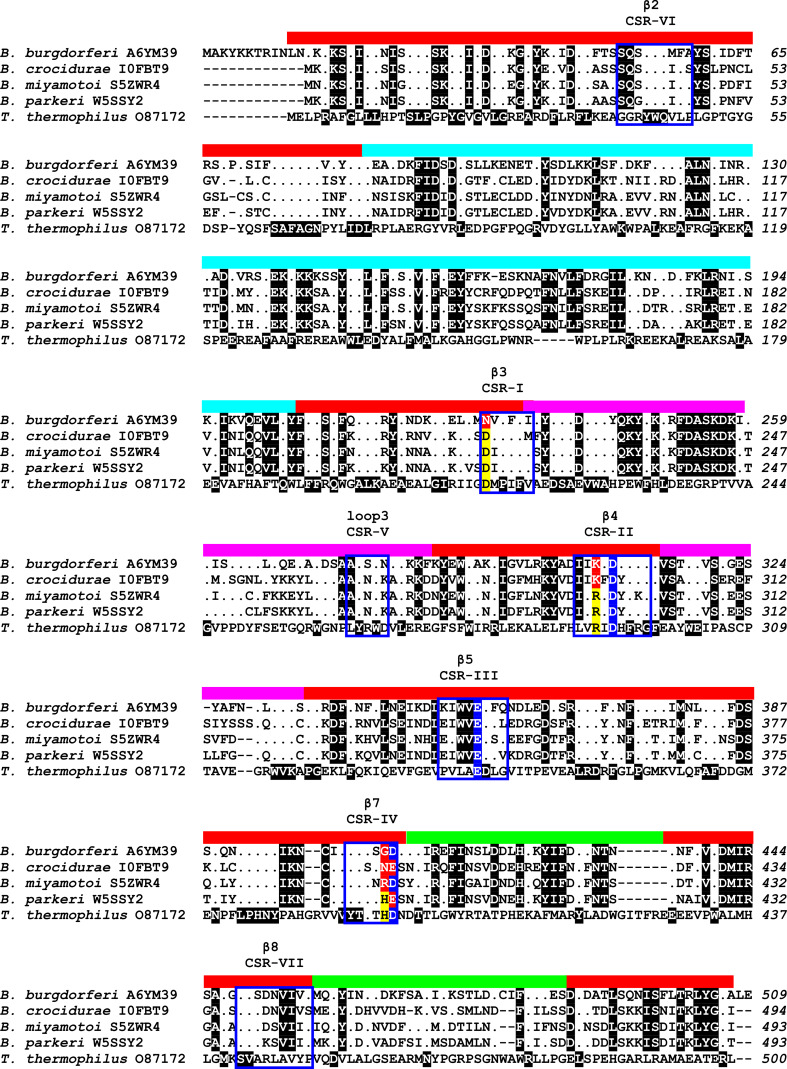
According to the most recent study focused on analysis of all available amylomaltase sequences from borreliae within the context of the entire family GH77 [122], 32 such sequences can be, in fact, divided into a few groups. Basically, there are only two major groups with respect to the presence/absence of the above-mentioned functional arginine, which—if naturally substituted—only lysine has been observed to replace it (Fig. 5c). It is the position of the Arg291 in the amylomaltase from Thermus aquaticus [43] mutated to Lys306 in the B. burgdorferi counterpart [129]. It should be pointed out that with regard to domain composition and length, all 32 amylomaltases from borreliae resemble typical bacterial GH77 amylomaltases represented by the enzyme from T. aquaticus. Nevertheless, some of those with the arginine to lysine mutation contain additional substitutions in several other well-conserved positions. On the other hand, some of those, possessing the original arginine, keep also those well-conserved positions (Fig. 6) characteristic for typical prokaryotic amylomaltases of non-borrelian origin [145]. Besides there are also some that exhibit an intermediary character. This phenomenon as a whole makes the amylomaltases from borreliae a unique evolutionary lineage in the family GH77 [122]. However, it should also be taken into account that until now of the 32 available GH77 amylomaltase sequences from borreliae, the MalQ from B. burgdorferi is the only one that has already been biochemically characterized and found enzymatically active [129, 146].
Fig. 6.
Amino acid sequence comparison of family GH77 amylomaltases from borreliae and Thermus aquaticus. The four amylomaltases from borrelian origin represent different subgroups within the genus Borrelia [122] exhibiting unique mutations in several important active-site positions with respect to a typical bacterial amylomaltase represented by the one from Thermus [129, 145]. The sequences were retrieved from the UniProt database [221] according to their accession numbers succeeding the name of the organism. The alignment was done using the program Clustal-Omega [230] available at the European Bioinformatics Institute’s web-site (http://www.ebi.ac.uk/). The unique borreliae-like positions (Asn228, Lys306 and Gly407) and the catalytic triad (Asp308, Glu355 and Asp408; B. burgdorferi amylomaltase numbering) are signified by yellow/red and blue highlighting, respectively. The positions in amylomaltases from borreliae identical to that from Thermus are shown as dots. The black highlighting in the T. aquaticus amylomaltase means that all four borrelian counterparts have the same residue in those positions. The positions that are signified by black highlighting in amylomaltases from borreliae are identical among them but different from those in the enzyme from Thermus. The seven CSRs known for the entire α-amylase clan GH-H are boxed and marked as CSR-I to CSR-VII with indicated well-accepted secondary structure elements [32]. The individual domains are indicated as a colored lane above the alignment blocks as follows: catalytic (β/α)8-barrel—red, subdomain B1—cyan, subdomain B2—magenta, subdomain B3—green
The family GH77 offers also other examples that are of interest from the evolutionary point of view, e.g., the DPE2 version of a typical plant DPE1 [42, 134, 135] possessing an insert of ~140 residues between catalytic nucleophile and proton donor [147–152] that have been found also among some bacteria [122], or a large group of additional bacterial amylomaltases represented by the enzyme from Escherichia coli [125] including also the well-characterized amylomaltase from Clostridium glutamicum [131, 153–155] that have at the N-terminus a separate carbohydrate-binding module-(CBM)-like domain [122, 142]. In any case it is evident that with regard to the family GH77 a gradual evolutionary transition has occurred among borreliae that can be characterized from the version of a typical bacterial Thermus-like amylomaltase to the version with progressively mutated functionally important conserved residues [122].
Other examples of interest
The main goal of this review article was to demonstrate some selected examples of remarkable evolutionary phenomena within the α-amylase clan GH-H. There are many other cases that would deserve to be mentioned, including also those seen in the second and smaller α-amylase family GH57. To mention at least a few, the non-exhaustive list could be as follows: (1) close similarity and relatedness between the family GH13 α-amylases from animals and actinomycetes [28, 74, 79] constituting a well-known group of chloride-dependent α-amylases [156, 157]; (2) existence of an intermediary group of α-amylases, classified in the subfamily GH13_36 [48], with a mixed enzyme specificity exhibiting simultaneous relatedness to various α-glucosidases and debranching enzymes [158–163] from the so-called subfamilies of oligo-1,6-glucosidase and neopullulanase [45, 46]; (3) presence of the so-called α-amylase-like homologues in the family GH57 [164–166] strikingly similar to α-amylases, but having incomplete GH57 catalytic machinery—the real α-amylases being more frequent among Archaea, whereas the α-amylase-like homologues are being found rather among Bacteria [164]; (4) the pronounced similarity between the α-amylase families GH57 and GH119 revealed by an in silico study suggesting that the members of the family GH119 share with those of the GH57 catalytic machinery, CSRs and fold of the catalytic domain [167]; (5) observation of a totally novel lineage or subfamily of glucansucrases within the circularly-permuted family GH70 [39–41, 168–170], exhibiting a close homology to the rest of the family but without any circular permutation of the catalytic TIM-barrel [171, 172]; (6) existence of various and evolutionarily independent starch/glycogen-binding domains classified as different CBM families [173–185] in, e.g., not typical amylolytic enzymes and proteins, such as glucan phosphatases—animal laforin and plant SEX-4 [186–197], AMP-activated protein kinase [198–201], genethonin-1 [202, 203], and starch synthases [204–206], glucan water dikinases [207–211] and lytic polysaccharide monooxygenases [212–214]; and (7) the so-called carbohydrate surface binding sites different from the distinct CBMs mentioned above and present in various carbohydrate-active enzymes, well represented just in the α-amylase family GH13 [69, 72, 215–219]—in the particular case of the barley α-amylase low pI isozyme AMY1 named as “a pair of sugar tongs” and a starch granule binding site, respectively [69, 218]. All these particular details make the α-amylases and the remaining enzyme specificities an attractive subject not only for evolutionary studies but also from the practical point of view for the approaches focused on their protein design. Even if known that some living organisms can survive without any sugars [220].
Conclusions
Due to a huge amount of accumulated sequence data, the questions concerning the structure, function and evolution of amylolytic enzymes have really become complicated, especially when they are ambitiously treated in an effort to reach answers to complex questions. This obvious trend can simply be illustrated by a few dozens of the α-amylase family GH13 members when it was created in 1991 versus more than 30 thousand sequences classified in the family nowadays. There is, however, not only the prevalent number of sequences available complicating the situation, i.e., our knowledge, but there is also the necessity to accept the changes in minds of scientists reflecting the continuously widening scope of the α-amylase family. Thus, for example, this family was originally established as a family of starch hydrolases and related enzymes active towards the α-1,4- and α-1,6-glucosidic linkages catalyzing either the hydrolysis (EC 3) or transglycosylation (EC 2). Currently, also other bonds are attacked, e.g., those in trehalose (α-1,1-) and sucrose (α-1,2-), the enzymatic repertoire of the family members has been expanded including the isomerases (EC 5). What might originally be unbelievable, even the non-enzymatic transport proteins (rBAT and 4F2hc), lacking in most cases the GH13 catalytic machinery, have been classified constituting their respective GH13 subfamilies. One of the ways that could lead to a consolidation of the knowledge may be to continue in looking for and revealing the features in amino acid sequences and structures that would reflect the exclusivity of smaller family groups (i.e., subfamilies) of either particular substrate specificity or from the point of view of taxonomy, or both, and confirming their roles experimentally.
Acknowledgments
SJ thanks for financial support from the Slovak Grant Agency VEGA (the grant No. 2/0150/14) and the Slovak Research and Development Agency (the contract No. LPP-0417-09). MG thanks for financial support from the Academician Stefan Schwarz Foundation of the Slovak Academy of Sciences.
Abbreviations
- CAZy
Carbohydrate-Active enZymes
- CBM
Carbohydrate-binding module
- CGTase
Cyclodextrin glucanotransferase
- CSR
Conserved sequence region
- DPE
Disproportionating enzyme
- GH
Glycoside hydrolase
- hcHATs
Heavy chains of the heteromeric amino acid transporters
References
- 1.Janecek S, Svensson B, MacGregor EA. α-Amylase—an enzyme specificity found in various families of glycoside hydrolases. Cell Mol Life Sci. 2014;71:1149–1170. doi: 10.1007/s00018-013-1388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacGregor EA. α-Amylase structure and activity. J Protein Chem. 1988;7:399–415. doi: 10.1007/BF01024888. [DOI] [PubMed] [Google Scholar]
- 3.Vihinen M, Mäntsälä P. Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol. 1989;24:329–418. doi: 10.3109/10409238909082556. [DOI] [PubMed] [Google Scholar]
- 4.Pandey A, Nigam P, Soccol CR, Soccol VT, Singh D, Mohan R. Advances in microbial amylases. Biotechnol Appl Biochem. 2000;31:135–152. doi: 10.1042/BA19990073. [DOI] [PubMed] [Google Scholar]
- 5.Søgaard M, Ji Abe, Martin-Eauclaire MF, Svensson B. α-Amylases: structure and function. Carbohydr Polym. 1993;21:137–146. doi: 10.1016/0144-8617(93)90008-R. [DOI] [Google Scholar]
- 6.Gupta R, Gigras P, Mohapatra H, Kumar Goswami V, Chauhan B. Microbial α-amylases: a biotechnological perspective. Process Biochem. 2003;38:1599–1616. doi: 10.1016/S0032-9592(03)00053-0. [DOI] [Google Scholar]
- 7.Svensson B. Regional distant sequence homology between amylases, α-glucosidases and transglucanosylases. FEBS Lett. 1988;230:72–76. doi: 10.1016/0014-5793(88)80644-6. [DOI] [PubMed] [Google Scholar]
- 8.MacGregor EA, Svensson B. A super-secondary structure predicted to be common to several α-1,4-D-glucan-cleaving enzymes. Biochem J. 1989;259:145–152. doi: 10.1042/bj2590145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jespersen HM, MacGregor EA, Sierks MR, Svensson B. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem J. 1991;280:51–55. doi: 10.1042/bj2800051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uitdehaag JC, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 11.Kimura K, Kataoka S, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the β-cyclodextrin glucanotransferase gene of alkalophilic Bacillus sp. strain 1011 and similarity of its amino acid sequence to those of α-amylases. J Bacteriol. 1987;169:4399–4402. doi: 10.1128/jb.169.9.4399-4402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janecek S, Svensson B, MacGregor EA. Characteristic differences in the primary structure allow discrimination of cyclodextrin glucanotransferases from α-amylases. Biochem J. 1995;305:685–686. doi: 10.1042/bj3050685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itkor P, Tsukagoshi N, Udaka S. Nucleotide sequence of the raw-starch-digesting amylase gene from Bacillus sp. B1018 and its strong homology to the cyclodextrin glucanotransferase genes. Biochem Biophys Res Commun. 1990;166:630–636. doi: 10.1016/0006-291X(90)90855-H. [DOI] [PubMed] [Google Scholar]
- 14.Bahl H, Burchhardt G, Spreinat A, Haeckel K, Wienecke A, Schmidt B, Antranikian G. α-Amylase of Clostridium thermosulfurogenes EM1: nucleotide sequence of the gene, processing of the enzyme, and comparison of other α-amylases. Appl Environ Microbiol. 1991;57:1554–1559. doi: 10.1128/aem.57.5.1554-1559.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del-Rio G, Morett E, Soberon X. Did cyclodextrin glycosyltransferases evolve from α-amylases? FEBS Lett. 1997;416:221–224. doi: 10.1016/S0014-5793(97)01192-7. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto A, Kimura K, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the maltohexaose-producing amylase gene from an alkalophilic Bacillus sp. #707 and structural similarity to liquefying type α-amylases. Biochem Biophys Res Commun. 1988;151:25–31. doi: 10.1016/0006-291X(88)90554-2. [DOI] [PubMed] [Google Scholar]
- 17.Imanaka T, Kuriki T. Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J Bacteriol. 1989;171:369–374. doi: 10.1128/jb.171.1.369-374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuriki T, Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus . J Gen Microbiol. 1989;135:1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- 19.Kuriki T, Takata H, Okada S, Imanaka T. Analysis of the active center of Bacillus stearothermophilus neopullulanase. J Bacteriol. 1991;173:6147–6152. doi: 10.1128/jb.173.19.6147-6152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1,4)- and α-(1,6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- 21.MacGregor EA. Relationships between structure and activity in the α-amylase family of starch-metabolising enzymes. Starch/Staerke. 1993;45:232–237. doi: 10.1002/star.19930450705. [DOI] [Google Scholar]
- 22.Jespersen HM, MacGregor EA, Henrissat B, Sierks MR, Svensson B. Starch- and glycogen-debranching and branching enzymes:prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- 23.Janecek S. Parallel β/α-barrels of α-amylase, cyclodextrin glycosyltransferase and oligo-1,6-glucosidase versus the barrel of β-amylase: evolutionary distance is a reflection of unrelated sequences. FEBS Lett. 1994;353:119–123. doi: 10.1016/0014-5793(94)01019-6. [DOI] [PubMed] [Google Scholar]
- 24.Svensson B. Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol Biol. 1994;25:141–157. doi: 10.1007/BF00023233. [DOI] [PubMed] [Google Scholar]
- 25.Aghajari N, Roth M, Haser R. Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic α-amylase. Biochemistry. 2002;41:4273–4280. doi: 10.1021/bi0160516. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol. 1986;23:355–360. doi: 10.1007/BF00257032. [DOI] [Google Scholar]
- 27.Janecek S. New conserved amino acid region of α-amylases in the third loop of their (β/α)8-barrel domains. Biochem J. 1992;288:1069–1070. doi: 10.1042/bj2881069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janecek S. Sequence similarities and evolutionary relationships of microbial, plant and animal α-amylases. Eur J Biochem. 1994;224:519–524. doi: 10.1111/j.1432-1033.1994.00519.x. [DOI] [PubMed] [Google Scholar]
- 29.Janecek S. Close evolutionary relatedness among functionally distantly related members of the (α/β)8-barrel glycosyl hydrolases suggested by the similarity of their fifth conserved sequence region. FEBS Lett. 1995;377:6–8. doi: 10.1016/0014-5793(95)01309-1. [DOI] [PubMed] [Google Scholar]
- 30.Janecek S. α-Amylase family: molecular biology and evolution. Prog Biophys Mol Biol. 1997;67:67–97. doi: 10.1016/S0079-6107(97)00015-1. [DOI] [PubMed] [Google Scholar]
- 31.MacGregor EA, Janecek S, Svensson B. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim Biophys Acta. 2001;1546:1–20. doi: 10.1016/S0167-4838(00)00302-2. [DOI] [PubMed] [Google Scholar]
- 32.Janecek S. How many conserved sequence regions are there in the α-amylase family? Biologia. 2002;57(Suppl 11):29–41. [Google Scholar]
- 33.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombard V, Ramulu GH, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucl Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henrissat B, Davies GJ. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/S0959-440X(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 39.MacGregor EA, Jespersen HM, Svensson B. A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 1996;378:263–266. doi: 10.1016/0014-5793(95)01428-4. [DOI] [PubMed] [Google Scholar]
- 40.Vujicic-Zagar A, Pijning T, Kralj S, Lopez CA, Eeuwema W, Dijkhuizen L, Dijkstra BW. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc Natl Acad Sci USA. 2010;107:21406–21411. doi: 10.1073/pnas.1007531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kralj S, Grijpstra P, van Leeuwen SS, Leemhuis H, Dobruchowska JM, van der Kaaij RM, Malik A, Oetari A, Kamerling JP, Dijkhuizen L. 4,6-α-Glucanotransferase, a novel enzyme that structurally and functionally provides an evolutionary link between glycoside hydrolase enzyme families 13 and 70. Appl Environ Microbiol. 2011;77:8154–8163. doi: 10.1128/AEM.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takaha T, Yanase M, Okada S, Smith SM. Disproportionating enzyme (4-α-glucanotransferase; EC 2.4.1.25) of potato. Purification, molecular cloning, and potential role in starch metabolism. J Biol Chem. 1993;268:1391–1396. [PubMed] [Google Scholar]
- 43.Terada Y, Fujii K, Takaha T, Okada S. Thermus aquaticus ATCC 33923 amylomaltase gene cloning and expression and enzyme characterization: production of cycloamylose. Appl Environ Microbiol. 1999;65:910–915. doi: 10.1128/aem.65.3.910-915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Przylas I, Tomoo K, Terada Y, Takaha T, Fujii K, Saenger W, Sträter N. Crystal structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans. J Mol Biol. 2000;296:873–886. doi: 10.1006/jmbi.1999.3503. [DOI] [PubMed] [Google Scholar]
- 45.Oslancova A, Janecek S. Oligo-1,6-glucosidase and neopullulanase enzyme subfamilies from the α-amylase family defined by the fifth conserved sequence region. Cell Mol Life Sci. 2002;59:1945–1959. doi: 10.1007/PL00012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies:towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 47.Lei Y, Peng H, Wang Y, Liu Y, Han F, Xiao Y, Gao Y. Preferential and rapid degradation of raw rice starch by an α-amylase of glycoside hydrolase subfamily GH13_37. Appl Microbiol Biotechnol. 2012;94:1577–1584. doi: 10.1007/s00253-012-4114-0. [DOI] [PubMed] [Google Scholar]
- 48.Majzlova K, Pukajova Z, Janecek S. Tracing the evolution of the α-amylase subfamily GH13_36 covering the amylolytic enzymes intermediate between oligo-1,6-glucosidases and neopullulanases. Carbohydr Res. 2013;367:48–57. doi: 10.1016/j.carres.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 49.Janecek S, Kuchtova A, Petrovicova S. A novel GH13 subfamily of α-amylases with a pair of tryptophans in the helix α3 of the catalytic TIM-barrel, the LPDlx signature in the conserved sequence region V and a conserved aromatic motif at the C-terminus. Biologia. 2015;70:1284–1294. doi: 10.1515/biolog-2015-0165. [DOI] [Google Scholar]
- 50.Gabrisko M, Janecek S. Looking for the ancestry of the heavy-chain subunits of heteromeric amino acid transporters rBAT and 4F2hc within the GH13 α-amylase family. FEBS J. 2009;276:7265–7278. doi: 10.1111/j.1742-4658.2009.07434.x. [DOI] [PubMed] [Google Scholar]
- 51.Dong G, Vieille C, Savchenko A, Zeikus JG. Cloning, sequencing, and expression of the gene encoding extracellular α-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3569–3576. doi: 10.1128/aem.63.9.3569-3576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen S, Vorgias CE, Antranikian G. Cloning, sequencing, characterization, and expression of an extracellular α-amylase from the hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli and Bacillus subtilis . J Biol Chem. 1997;272:16335–16342. doi: 10.1074/jbc.272.26.16335. [DOI] [PubMed] [Google Scholar]
- 53.Jones RA, Jermiin LS, Easteal S, Patel BK, Beacham IR. Amylase and 16S rRNA genes from a hyperthermophilic archaebacterium. J Appl Microbiol. 1999;86:93–107. doi: 10.1046/j.1365-2672.1999.00642.x. [DOI] [PubMed] [Google Scholar]
- 54.Frillingos S, Linden A, Niehaus F, Vargas C, Nieto JJ, Ventosa A, Antranikian G, Drainas C. Cloning and expression of α-amylase from the hyperthermophilic archaeon Pyrococcus woesei in the moderately halophilic bacterium Halomonas elongata . J Appl Microbiol. 2000;88:495–503. doi: 10.1046/j.1365-2672.2000.00988.x. [DOI] [PubMed] [Google Scholar]
- 55.Leveque E, Haye B, Belarbi A. Cloning and expression of an α-amylase encoding gene from the hyperthermophilic archaebacterium Thermococcus hydrothermalis and biochemical characterisation of the recombinant enzyme. FEMS Microbiol Lett. 2000;186:67–71. doi: 10.1016/S0378-1097(00)00117-8. [DOI] [PubMed] [Google Scholar]
- 56.Rogers JC, Milliman C. Isolation and sequence analysis of a barley α-amylase cDNA clone. J Biol Chem. 1983;258:8169–8174. [PubMed] [Google Scholar]
- 57.Rogers JC. Two barley α-amylase gene families are regulated differently in aleurone cells. J Biol Chem. 1985;260:3731–3738. [PubMed] [Google Scholar]
- 58.Baulcombe DC, Huttly AK, Martienssen RA, Barker RF, Jarvis MG. A novel wheat α-amylase gene (α-Amy3) Mol Gen Genet. 1987;209:33–40. doi: 10.1007/BF00329833. [DOI] [PubMed] [Google Scholar]
- 59.Huang N, Sutliff TD, Litts JC, Rodriguez RL. Plant Mol Biol. 1990;14:655–668. doi: 10.1007/BF00016499. [DOI] [PubMed] [Google Scholar]
- 60.Young TE, DeMason DA, Close TJ. Cloning of an α-amylase cDNA from aleurone tissue of germinating maize seed. Plant Physiol. 1994;105:759–760. doi: 10.1104/pp.105.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kadziola A, Abe J, Svensson B, Haser R. Crystal and molecular structure of barley α-amylase. J Mol Biol. 1994;239:104–121. doi: 10.1006/jmbi.1994.1354. [DOI] [PubMed] [Google Scholar]
- 62.Kadziola A, Søgaard M, Svensson B, Haser R. Molecular structure of a barley α-amylase-inhibitor complex:implications for starch binding and catalysis. J Mol Biol. 1998;278:205–217. doi: 10.1006/jmbi.1998.1683. [DOI] [PubMed] [Google Scholar]
- 63.Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984;95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 64.Swift HJ, Brady L, Derewenda ZS, Dodson EJ, Dodson GG, Turkenburg JP, Wilkinson AJ. Structure and molecular model refinement of Aspergillus oryzae (TAKA) α-amylase: an application of the simulated-annealing method. Acta Crystallogr B. 1991;47:535–544. doi: 10.1107/S0108768191001970. [DOI] [PubMed] [Google Scholar]
- 65.Boel E, Brady L, Brzozowski AM, Derewenda Z, Dodson GG, Jensen VJ, Petersen SB, Swift H, Thim L, Woldike HF. Calcium binding in α-amylases: an X-ray diffraction study at 2.1-Å resolution of two enzymes from Aspergillus . Biochemistry. 1990;29:6244–6249. doi: 10.1021/bi00478a019. [DOI] [PubMed] [Google Scholar]
- 66.Brady RL, Brzozowski AM, Derewenda ZS, Dodson EJ, Dodson GG. Solution of the structure of Aspergillus niger acid α-amylase by combined molecular replacement and multiple isomorphous replacement methods. Acta Crystallogr B. 1991;47:527–535. doi: 10.1107/S0108768191001908. [DOI] [PubMed] [Google Scholar]
- 67.Buisson G, Duee E, Haser R, Payan F. Three dimensional structure of porcine pancreatic α-amylase at 2.9 Å resolution. Role of calcium in structure and activity. EMBO J. 1987;6:3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian M, Haser R, Payan F. Structure and molecular model refinement of pig pancreatic α-amylase at 2.1 Å resolution. J Mol Biol. 1993;231:785–799. doi: 10.1006/jmbi.1993.1326. [DOI] [PubMed] [Google Scholar]
- 69.Robert X, Haser R, Gottschalk TE, Ratajczak F, Driguez H, Svensson B, Aghajari N. The structure of barley α-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs. Structure. 2003;11:973–984. doi: 10.1016/S0969-2126(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 70.Robert X, Haser R, Mori H, Svensson B, Aghajari N. Oligosaccharide binding to barley α-amylase 1. J Biol Chem. 2005;280:32968–32978. doi: 10.1074/jbc.M505515200. [DOI] [PubMed] [Google Scholar]
- 71.Ochiai A, Sugai H, Harada K, Tanaka S, Ishiyama Y, Ito K, Tanaka T, Uchiumi T, Taniguchi M, Mitsui T. Crystal structure of α-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability. Biosci Biotechnol Biochem. 2014;78:989–997. doi: 10.1080/09168451.2014.917261. [DOI] [PubMed] [Google Scholar]
- 72.Linden A, Mayans O, Meyer-Klaucke W, Antranikian G, Wilmanns M. Differential regulation of a hyperthermophilic α-amylase with a novel (Ca, Zn) two-metal center by zinc. J Biol Chem. 2003;278:9875–9884. doi: 10.1074/jbc.M211339200. [DOI] [PubMed] [Google Scholar]
- 73.Pujadas G, Palau J. Evolution of α-amylases:architectural features and key residues in the stabilization of the (β/α)8 scaffold. Mol Biol Evol. 2001;18:38–54. doi: 10.1093/oxfordjournals.molbev.a003718. [DOI] [PubMed] [Google Scholar]
- 74.Da Lage JL, Feller G, Janecek S. Horizontal gene transfer from Eukarya to bacteria and domain shuffling: the α-amylase model. Cell Mol Life Sci. 2004;61:97–109. doi: 10.1007/s00018-003-3334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Kaaij RM, Janecek S, van der Maarel MJ, Dijkhuizen L. Phylogenetic and biochemical characterization of a novel cluster of intracellular fungal α-amylase enzymes. Microbiology. 2007;153:4003–4015. doi: 10.1099/mic.0.2007/008607-0. [DOI] [PubMed] [Google Scholar]
- 76.Godany A, Majzlova K, Horvathova V, Vidova B, Janecek S. Tyrosine 39 of GH13 α-amylase from Thermococcus hydrothermalis contributes to its thermostability. Biologia. 2010;65:408–415. doi: 10.2478/s11756-010-0030-x. [DOI] [Google Scholar]
- 77.Hostinova E, Janecek S, Gasperik J. Gene sequence, bioinformatics and enzymatic characterization of α-amylase from Saccharomycopsis fibuligera KZ. Protein J. 2010;29:355–364. doi: 10.1007/s10930-010-9260-6. [DOI] [PubMed] [Google Scholar]
- 78.Puspasari F, Radjasa O, Noer A, Nurachman Z, Syah Y, van der Maarel M, Dijkhuizen L, Janecek S, Natalia D. Raw starch degrading α-amylase from Bacillus aquimaris MKSC 6.2: isolation and expression of the gene, bioinformatics and biochemical characterization of the recombinant enzyme. J Appl Microbiol. 2013;114:108–120. doi: 10.1111/jam.12025. [DOI] [PubMed] [Google Scholar]
- 79.Janecek S, Leveque E, Belarbi A, Haye B. Close evolutionary relatedness of α-amylases from Archaea and plants. J Mol Evol. 1999;48:421–426. doi: 10.1007/PL00006486. [DOI] [PubMed] [Google Scholar]
- 80.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 81.Li C, Du M, Cheng B, Wang L, Liu X, Ma C, Yang C, Xu P. Close relationship of a novel Flavobacteriaceae α-amylase with archaeal α-amylases and good potentials for industrial applications. Biotechnol Biofuels. 2014;7:18. doi: 10.1186/1754-6834-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linden A, Wilmanns M. Adaptation of class-13 α-amylases to diverse living conditions. ChemBioChem. 2004;5:231–239. doi: 10.1002/cbic.200300734. [DOI] [PubMed] [Google Scholar]
- 83.Lim JK, Lee HS, Kim YJ, Bae SS, Jeon JH, Kang SG, Lee JH. Critical factors to high thermostability of an α-amylase from hyperthermophilic archaeon Thermococcus onnurineus NA1. J Microbiol Biotechnol. 2007;17:1242–1248. [PubMed] [Google Scholar]
- 84.Kobayashi T, Kanai H, Hayashi T, Akiba T, Akaboshi R, Horikoshi K. Haloalkaliphilic maltotriose-forming α-amylase from the archaebacterium Natronococcus sp. strain Ah-36. J Bacteriol. 1992;174:3439–3444. doi: 10.1128/jb.174.11.3439-3444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi T, Kanai H, Aono R, Horikoshi K, Kudo T. Cloning, expression, and nucleotide sequence of the α-amylase gene from the haloalkaliphilic archaeon Natronococcus sp. strain Ah-36. J Bacteriol. 1994;176:5131–5134. doi: 10.1128/jb.176.16.5131-5134.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onodera M, Yatsunami R, Tsukimura W, Fukui T, Nakasone K, Takashina T, Nakamura S. Gene analysis, expression, and characterization of an intracellular α-amylase from the extremely halophilic archaeon Haloarcula japonica . Biosci Biotechnol Biochem. 2013;77:281–288. doi: 10.1271/bbb.120693. [DOI] [PubMed] [Google Scholar]
- 87.Zorgani MA, Patron K, Desvaux M. New insight in the structural features of haloadaptation in α-amylases from halophilic Archaea following homology modeling strategy: folded and stable conformation maintained through low hydrophobicity and highly negative charged surface. J Comput Aided Mol Des. 2014;28:721–734. doi: 10.1007/s10822-014-9754-y. [DOI] [PubMed] [Google Scholar]
- 88.Hutcheon GW, Vasisht N, Bolhuis A. Characterisation of a highly stable α-amylase from the halophilic archaeon Haloarcula hispanica . Extremophiles. 2005;9:487–495. doi: 10.1007/s00792-005-0471-2. [DOI] [PubMed] [Google Scholar]
- 89.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 90.Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. Crystal structure of Bacillus stearothermophilus α-amylase: possible factors determining the thermostability. J Biochem. 2001;129:461–468. doi: 10.1093/oxfordjournals.jbchem.a002878. [DOI] [PubMed] [Google Scholar]
- 91.Wells RG, Hediger MA. Cloning of a rat kidney cDNA that stimulates dibasic and neutral amino acid transport and has sequence similarity to glucosidases. Proc Natl Acad Sci USA. 1992;89:5596–5600. doi: 10.1073/pnas.89.12.5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bertran J, Werner A, Moore ML, Stange G, Markovich D, Biber J, Testar X, Zorzano A, Palacin M, Murer H. Expression cloning of a cDNA from rabbit kidney cortex that induces a single transport system for cystine and dibasic and neutrl amino acids. Proc Natl Acad Sci USA. 1992;89:5601–5605. doi: 10.1073/pnas.89.12.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuriki T, Imanaka T. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87:557–565. doi: 10.1016/S1389-1723(99)80114-5. [DOI] [PubMed] [Google Scholar]
- 94.Kuriki T, Takata H, Yanase M, Ohdan K, Fujii K, Terada Y, Takaha T, Hondoh H, Matsuura Y, Imanaka T. The concept of the α-amylase family: a rational tool for interconverting glucanohydrolases/glucanotransferases, and their specificities. J Appl Glycosci. 2006;53:155–161. doi: 10.5458/jag.53.155. [DOI] [Google Scholar]
- 95.Janecek S, Svensson B, Henrissat B. Domain evolution in the α-amylase family. J Mol Evol. 1997;45:322–331. doi: 10.1007/PL00006236. [DOI] [PubMed] [Google Scholar]
- 96.Watanabe K, Hata Y, Kizaki H, Katsube Y, Suzuki Y. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J Mol Biol. 1997;269:142–153. doi: 10.1006/jmbi.1997.1018. [DOI] [PubMed] [Google Scholar]
- 97.Zhang D, Li N, Lok SM, Zhang LH, Swaminathan K. Isomaltulose synthase (PalI) of Klebsiella sp. LX3. Crystal structure and implication of mechanism. J Biol Chem. 2003;278:35428–35434. doi: 10.1074/jbc.M302616200. [DOI] [PubMed] [Google Scholar]
- 98.Hondoh H, Saburi W, Mori H, Okuyama M, Nakada T, Matsuura Y, Kimura A. Substrate recognition mechanism of α-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans . J Mol Biol. 2008;378:913–922. doi: 10.1016/j.jmb.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 99.Shirai T, Hung VS, Morinaka K, Kobayashi T, Ito S. Crystal structure of GH13 α-glucosidase GSJ from one of the deepest sea bacteria. Proteins. 2008;73:126–133. doi: 10.1002/prot.22044. [DOI] [PubMed] [Google Scholar]
- 100.Kim JS, Cha SS, Kim HJ, Kim TJ, Ha NC, Oh ST, Cho HS, Cho MJ, Kim MJ, Lee HS, Kim JW, Choi KY, Park KH, Oh BH. Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J Biol Chem. 1999;274:26279–26286. doi: 10.1074/jbc.274.37.26279. [DOI] [PubMed] [Google Scholar]
- 101.Lee HS, Kim MS, Cho HS, Kim JI, Kim TJ, Choi JH, Park C, Lee HS, Oh BH, Park KH. Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J Biol Chem. 2002;277:21891–21897. doi: 10.1074/jbc.M201623200. [DOI] [PubMed] [Google Scholar]
- 102.Hondoh H, Kuriki T, Matsuura Y. Three-dimensional structure and substrate binding of Bacillus stearothermophilus neopullulanase. J Mol Biol. 2003;326:177–188. doi: 10.1016/S0022-2836(02)01402-X. [DOI] [PubMed] [Google Scholar]
- 103.Fort J, de la Ballina LR, Burghardt HE, Ferrer-Costa C, Turnay J, Ferrer-Orta C, Uson I, Zorzano A, Fernandez-Recio J, Orozco M, Lizarbe MA, Fita I, Palacin M. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J Biol Chem. 2007;282:31444–31452. doi: 10.1074/jbc.M704524200. [DOI] [PubMed] [Google Scholar]
- 104.Janecek S. Proteins without enzymatic function with sequence relatedness to the α-amylase family. Trends Glycosci Glycotechnol. 2000;12:363–371. doi: 10.4052/tigg.12.363. [DOI] [Google Scholar]
- 105.Broer S, Wagner CA. Structure-function relationships of heterodimeric amino acid transporters. Cell Biochem Biophys. 2002;36:155–168. doi: 10.1385/CBB:36:2-3:155. [DOI] [PubMed] [Google Scholar]
- 106.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflug Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 107.Palacin M, Nunes V, Font-Llitjos M, Jimenez-Vidal M, Fort J, Gasol E, Pineda M, Feliubadalo L, Chillaron J, Zorzano A. The genetics of heteromeric amino acid transporters. Physiology. 2005;20:112–124. doi: 10.1152/physiol.00051.2004. [DOI] [PubMed] [Google Scholar]
- 108.Chillaron J, Roca R, Valencia A, Zorzano A, Palacin M. Heteromeric amino acid transporters: biochemistry, genetics, and physiology. Am J Physiol Renal Physiol. 2001;281:995–1018. doi: 10.1152/ajprenal.2001.281.6.F995. [DOI] [PubMed] [Google Scholar]
- 109.Chillaron J, Font-Llitjos M, Fort J, Zorzano A, Goldfarb DS, Nunes V, Palacin M. Pathophysiology and treatment of cystinuria. Nat Rev Nephrol. 2010;6:424–434. doi: 10.1038/nrneph.2010.69. [DOI] [PubMed] [Google Scholar]
- 110.Bertran J, Werner A, Chillaron J, Nunes V, Biber J, Testar X, Zorzano A, Estivill X, Murer H, Palacin M. Expression cloning of a human renal cDNA that induces high affinity transport of L-cystine shared with dibasic amino acids in Xenopus oocytes. J Biol Chem. 1993;268:14842–14849. [PubMed] [Google Scholar]
- 111.Lee WS, Wells RG, Sabbag RV, Mohandas TK, Hediger MA. Cloning and chromosomal localization of a human kidney cDNA involved in cystine, dibasic, and neutral amino acid transport. J Clin Invest. 1993;91:1959–1963. doi: 10.1172/JCI116415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reig N, Chillaron J, Bartoccioni P, Fernandez E, Bendahan A, Zorzano A, Kanner B, Palacin M, Bertran J. The light subunit of system b0, + is fully functional in the absence of the heavy subunit. EMBO J. 2002;21:4906–4914. doi: 10.1093/emboj/cdf500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fernandez E, Jimenez-Vidal M, Calvo M, Zorzano A, Tebar F, Palacin M, Chillaron J. The structural and functional units of heteromeric amino acid transporters. The heavy subunit rBAT dictates oligomerization of the heteromeric amino acid transporters. J Biol Chem. 2006;281:26552–26556. doi: 10.1074/jbc.M604049200. [DOI] [PubMed] [Google Scholar]
- 114.Rosell A, Meury M, Alvarez-Marimon E, Costa M, Perez-Cano L, Zorzano A, Fernandez-Recio J, Palacin M, Fotiadis D. Structural bases for the interaction and stabilization of the human amino acid transporter LAT2 with its ancillary protein 4F2hc. Proc Natl Acad Sci USA. 2014;111:2966–2971. doi: 10.1073/pnas.1323779111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park KH, Kim TJ, Cheong TK, Kim JW, Oh BH, Svensson B. Structure, specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochim Biophys Acta. 2000;1478:165–185. doi: 10.1016/S0167-4838(00)00041-8. [DOI] [PubMed] [Google Scholar]
- 116.Jin X, Aimanova K, Ross LS, Gill SS. Identification, functional characterization and expression of a LAT type amino acid transporter from the mosquito Aedes aegypti . Insect Biochem Mol Biol. 2003;33:815–827. doi: 10.1016/S0965-1748(03)00081-X. [DOI] [PubMed] [Google Scholar]
- 117.Reynolds B, Roversi P, Laynes R, Kazi S, Boyd CA, Goberdhan DC. Drosophila expresses a CD98 transporter with an evolutionarily conserved structure and amino acid-transport properties. Biochem J. 2009;420:363–372. doi: 10.1042/BJ20082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Veljkovic E, Stasiuk S, Skelly PJ, Shoemaker CB, Verrey F. Functional characterization of Caenorhabditis elegans heteromeric amino acid transporters. J Biol Chem. 2004;279:7655–7662. doi: 10.1074/jbc.M309528200. [DOI] [PubMed] [Google Scholar]
- 119.Krautz-Peterson G, Camargo S, Huggel K, Verrey F, Shoemaker CB, Skelly PJ. Amino acid transport in schistosomes: characterization of the permease heavy chain SPRM1hc. J Biol Chem. 2007;282:21767–21775. doi: 10.1074/jbc.M703512200. [DOI] [PubMed] [Google Scholar]
- 120.Gabrisko M. Evolutionary history of eukaryotic α-glucosidases from the α-amylase family. J Mol Evol. 2013;76:129–145. doi: 10.1007/s00239-013-9545-4. [DOI] [PubMed] [Google Scholar]
- 121.Verrey F, Jack DL, Paulsen IT, Saier MH, Jr, Pfeiffer R. New glycoprotein-associated amino acid transporters. J Membr Biol. 1999;172:181–192. doi: 10.1007/s002329900595. [DOI] [PubMed] [Google Scholar]
- 122.Kuchtova A, Janecek S. In silico analysis of family GH77 with focus on amylomaltases from borreliae and disproportionating enzymes DPE2 from plants and bacteria. Biochim Biophys Acta. 2015;1854:1260–1268. doi: 10.1016/j.bbapap.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 123.Smith AM, Zeeman SC, Thorneycroft D, Smith SM. Starch mobilization in leaves. J Exp Bot. 2003;54:577–583. doi: 10.1093/jxb/erg036. [DOI] [PubMed] [Google Scholar]
- 124.Kaper T, van der Maarel MJ, Euverink GJ, Dijkhuizen L. Exploring and exploiting starch-modifying amylomaltases from thermophiles. Biochem Soc Trans. 2004;32:279–282. doi: 10.1042/bst0320279. [DOI] [PubMed] [Google Scholar]
- 125.Pugsley AP, Dubreuil C. Molecular characterization of malQ, the structural gene for the Escherichia coli enzyme amylomaltase. Mol Microbiol. 1988;2:473–479. doi: 10.1111/j.1365-2958.1988.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 126.Goda SK, Eissa O, Akhtar M, Minton NP. Molecular analysis of a Clostridium butyricum NCIMB 7423 gene encoding 4-α-glucanotransferase and characterization of the recombinant enzyme produced in Escherichia coli . Microbiology. 1997;143:3287–3294. doi: 10.1099/00221287-143-10-3287. [DOI] [PubMed] [Google Scholar]
- 127.Bhuiyan SH, Kitaoka M, Hayashi K. A cycloamylose-forming hyperthermostable 4-α-glucanotransferase of Aquifex aeolicus expressed in Escherichia coli . J Mol Catal B Enzym. 2003;22:45–53. doi: 10.1016/S1381-1177(03)00005-5. [DOI] [Google Scholar]
- 128.Kaper T, Talik B, Ettema TJ, Bos H, van der Maarel MJ, Dijkhuizen L. Amylomaltase of Pyrobaculum aerophilum IM2 produces thermoreversible starch gels. Appl Environ Microbiol. 2005;71:5098–5106. doi: 10.1128/AEM.71.9.5098-5106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Godany A, Vidova B, Janecek S. The unique glycoside hydrolase family 77 amylomaltase from Borrelia burgdorferi with only catalytic triad conserved. FEMS Microbiol Lett. 2008;284:84–91. doi: 10.1111/j.1574-6968.2008.01191.x. [DOI] [PubMed] [Google Scholar]
- 130.Lee BH, Oh DK, Yoo SH. Characterization of 4-α-glucanotransferase from Synechocystis sp. PCC 6803 and its application to various corn starches. N Biotechnol. 2009;26:29–36. doi: 10.1016/j.nbt.2009.06.981. [DOI] [PubMed] [Google Scholar]
- 131.Srisimarat W, Powviriyakul A, Kaulpiboon J, Krusong K, Zimmermann W, Pongsawasdi P. A novel amylomaltase from Corynebacterium glutamicum and analysis of the large-ring cyclodextrin products. J Incl Phenom Macrocycl Chem. 2011;70:369–375. doi: 10.1007/s10847-010-9890-5. [DOI] [Google Scholar]
- 132.Hwang S, Choi KH, Kim J, Cha J. Biochemical characterization of 4-α-glucanotransferase from Saccharophagus degradans 2-40 and its potential role in glycogen degradation. FEMS Microbiol Lett. 2013;344:145–151. doi: 10.1111/1574-6968.12167. [DOI] [PubMed] [Google Scholar]
- 133.Sawasdee S, Rudeekulthamrong P, Zimmermann W, Murakami S, Pongsawasdi P, Kaulpiboo J. Direct cloning of gene encoding a novel amylomaltase from soil bacterial DNA for large-ring cyclodextrin production. Appl Biochem Microbiol. 2014;50:17–24. doi: 10.1134/S000368381306015X. [DOI] [PubMed] [Google Scholar]
- 134.Wattebled F, Ral JP, Dauvillee D, Myers AM, James MG, Schlichting R, Giersch C, Ball SG, D’Hulst C. STA11, a Chlamydomonas reinhardtii locus required for normal starch granule biogenesis, encodes disproportionating enzyme. Further evidence for a function of α-1,4 glucanotransferases during starch granule biosynthesis in green algae. Plant Physiol. 2003;132:137–145. doi: 10.1104/pp.102.016527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bresolin NS, Li Z, Kosar-Hashemi B, Tetlow IJ, Chatterjee M, Rahman S, Morell MK, Howitt CA. Characterisation of disproportionating enzyme from wheat endosperm. Planta. 2006;224:20–31. doi: 10.1007/s00425-005-0187-7. [DOI] [PubMed] [Google Scholar]
- 136.van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol. 2002;94:137–155. doi: 10.1016/S0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 137.van der Maarel MJ, Leemhuis H. Starch modification with microbial α-glucanotransferase enzymes. Carbohydr Polym. 2013;93:116–121. doi: 10.1016/j.carbpol.2012.01.065. [DOI] [PubMed] [Google Scholar]
- 138.Ahmad N, Mehboob S, Rashid N. Starch-processing enzymes– emphasis on thermostable 4-α-glucanotransferases. Biologia. 2015;70:709–725. doi: 10.1515/biolog-2015-0087. [DOI] [Google Scholar]
- 139.Przylas I, Terada Y, Fujii K, Takaha T, Saenger W, Sträter N. X-ray structure of acarbose bound to amylomaltase from Thermus aquaticus. Implications for the synthesis of large cyclic glucans. Eur J Biochem. 2000;267:6903–6913. doi: 10.1046/j.1432-1033.2000.01790.x. [DOI] [PubMed] [Google Scholar]
- 140.Barends TR, Bultema JB, Kaper T, van der Maarel MJ, Dijkhuizen L, Dijkstra BW. Three-way stabilization of the covalent intermediate in amylomaltase, an α-amylase-like transglycosylase. J Biol Chem. 2007;282:17242–17249. doi: 10.1074/jbc.M701444200. [DOI] [PubMed] [Google Scholar]
- 141.Jung JH, Jung TY, Seo DH, Yoon SM, Choi HC, Park BC, Park CS, Woo EJ. Structural and functional analysis of substrate recognition by the 250s loop in amylomaltase from Thermus brockianus . Proteins. 2011;79:633–644. doi: 10.1002/prot.22911. [DOI] [PubMed] [Google Scholar]
- 142.Weiss SC, Skerra A, Schiefner A. Structural basis for the interconversion of maltodextrins by MalQ, the amylomaltase of Escherichia coli . J Biol Chem. 2015;290:21352–21364. doi: 10.1074/jbc.M115.667337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O’Neill EC, Stevenson CE, Tantanarat K, Latousakis D, Donaldson MI, Rejzek M, Nepogodiev SA, Limpaseni T, Field RA, Lawson DM. Structural dissection of the maltodextrin disproportionation cycle of the Arabidopsis plastidial enzyme DPE1. J Biol Chem. 2015;290:29834–29853. doi: 10.1074/jbc.M115.682245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi . Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 145.Machovic M, Janecek S. The invariant residues in the α-amylase family: just the catalytic triad. Biologia. 2003;58:1127–1132. [Google Scholar]
- 146.Hoon-Hanks LL, Morton EA, Lybecker MC, Battisti JM, Samuels DS, Drecktrah D. Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS Immunol Med Microbiol. 2012;66:157–165. doi: 10.1111/j.1574-695X.2012.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lloyd JR, Blennow A, Burhenne K, Kossmann J. Repression of a novel isoform of disproportionating enzyme (stDPE2) in potato leads to inhibition of starch degradation in leaves but not tubers stored at low temperature. Plant Physiol. 2004;134:1347–1354. doi: 10.1104/pp.103.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lütken H, Lloyd JR, Glaring MA, Baunsgaard L, Laursen KH, Haldrup A, Kossmann J, Blennow A. Repression of both isoforms of disproportionating enzyme leads to higher malto-oligosaccharide content and reduced growth in potato. Planta. 2010;232:1127–1139. doi: 10.1007/s00425-010-1245-3. [DOI] [PubMed] [Google Scholar]
- 149.Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 2004;37:853–863. doi: 10.1111/j.1365-313X.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- 150.Lu Y, Sharkey TD. The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta. 2004;218:466–473. doi: 10.1007/s00425-003-1127-z. [DOI] [PubMed] [Google Scholar]
- 151.Steichen JM, Petty RV, Sharkey TD. Domain characterization of a 4-α-glucanotransferase essential for maltose metabolism in photosynthetic leaves. J Biol Chem. 2008;283:20797–20804. doi: 10.1074/jbc.M803051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ruzanski C, Smirnova J, Rejzek M, Cockburn D, Pedersen HL, Pike M, Willats WG, Svensson B, Steup M, Ebenhöh O, Smith AM, Field RA. A bacterial glucanotransferase can replace the complex maltose metabolism required for starch to sucrose conversion in leaves at night. J Biol Chem. 2013;288:28581–28598. doi: 10.1074/jbc.M113.497867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Srisimarat W, Murakami S, Pongsawasdi P, Krusong K. Crystallization and preliminary X-ray crystallographic analysis of the amylomaltase from Corynebacterium glutamicum . Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2013;69:1004–1006. doi: 10.1107/S1744309113020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Srisimarat W, Kaulpiboon J, Krusong K, Zimmermann W, Pongsawasdi P. Altered large-ring cyclodextrin product profile due to a mutation at Tyr-172 in the amylomaltase of Corynebacterium glutamicum . Appl Environ Microbiol. 2012;78:7223–7228. doi: 10.1128/AEM.01366-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rachadech W, Nimpiboon P, Naumthong W, Nakapong S, Krusong K, Pongsawasdi P. Identification of essential tryptophan in amylomaltase from Corynebacterium glutamicum . Int J Biol Macromol. 2015;76:230–235. doi: 10.1016/j.ijbiomac.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 156.D’Amico S, Gerday C, Feller G. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene. 2000;253:95–105. doi: 10.1016/S0378-1119(00)00229-8. [DOI] [PubMed] [Google Scholar]
- 157.Da Lage JL, Danchin EG, Casane D. Where do animal α-amylases come from? An interkingdom trip. FEBS Lett. 2007;581:3927–3935. doi: 10.1016/j.febslet.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 158.Sivakumar N, Li N, Tang JW, Patel BK, Swaminathan K. Crystal structure of AmyA lacks acidic surface and provide insights into protein stability at poly-extreme condition. FEBS Lett. 2006;580:2646–2652. doi: 10.1016/j.febslet.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 159.Møller MS, Fredslund F, Majumder A, Nakai H, Poulsen JC, Lo Leggio L, Svensson B, Abou Hachem M. Enzymology and structure of the GH13_31 glucan 1,6-α-glucosidase that confers isomaltooligosaccharide utilization in the probiotic Lactobacillus acidophilus NCFM. J Bacteriol. 2012;194:4249–4259. doi: 10.1128/JB.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kobayashi M, Saburi W, Nakatsuka D, Hondoh H, Kato K, Okuyama M, Mori H, Kimura A, Yao M. Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase. FEBS Lett. 2015;589:484–489. doi: 10.1016/j.febslet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 161.Saburi W, Rachi-Otsuka H, Hondoh H, Okuyama M, Mori H, Kimura A. Structural elements responsible for the glucosidic linkage-selectivity of a glycoside hydrolase family 13 exo-glucosidase. FEBS Lett. 2015;589:865–869. doi: 10.1016/j.febslet.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 162.Møller MS, Henriksen A, Svensson B. Structure and function of α-glucan debranching enzymes. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Okuyama M, Saburi W, Mori H, Kimura A. α-Glucosidases and α-1,4-glucan lyases: structures, functions, and physiological actions. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Janecek S, Blesak K. Sequence-structural features and evolutionary relationships of family GH57 α-amylases and their putative α-amylase-like homologues. Protein J. 2011;30:429–435. doi: 10.1007/s10930-011-9348-7. [DOI] [PubMed] [Google Scholar]
- 165.Blesak K, Janecek S. Sequence fingerprints of enzyme specificities from the glycoside hydrolase family GH57. Extremophiles. 2012;16:497–506. doi: 10.1007/s00792-012-0449-9. [DOI] [PubMed] [Google Scholar]
- 166.Blesak K, Janecek S. Two potentially novel amylolytic enzyme specificities in the prokaryotic glycoside hydrolase α-amylase family GH57. Microbiology. 2013;159:2584–2593. doi: 10.1099/mic.0.071084-0. [DOI] [PubMed] [Google Scholar]
- 167.Janecek S, Kuchtova A. In silico identification of catalytic residues and domain fold of the family GH119 sharing the catalytic machinery with the α-amylase family GH57. FEBS Lett. 2012;586:3360–3366. doi: 10.1016/j.febslet.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 168.Passerini D, Vuillemin M, Ufarte L, Morel S, Loux V, Fontagné-Faucher C, Monsan P, Remaud-Simeon M, Moulis C. Inventory of the GH70 enzymes encoded by Leuconostoc citreum NRRL B-1299—identification of three novel α-transglucosylases. FEBS J. 2015;282:2115–2130. doi: 10.1111/febs.13261. [DOI] [PubMed] [Google Scholar]
- 169.Moulis C, André I, Remaud-Simeon M. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Meng X, Gangoiti J, Bai Y, Pijning T, Van Leeuwen SS, Dijkhuizen L. Structure-function relationships of family GH70 glucansucrase and 4,6-α-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gangoiti J, Pijning T, Dijkhuizen L. Biochemical characterization of the Exiguobacterium sibiricum 255-15 GtfC enzyme representing a novel glycoside hydrolase 70 subfamily of 4,6-α-glucanotransferase enzymes. Appl Environ Microbiol. 2016;82:756–766. doi: 10.1128/AEM.03420-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gangoiti J, van Leeuwen SS, Vafiadi C, Dijkhuizen L. The Gram-negative bacterium Azotobacter chroococcum NCIMB 8003 employs a new glycoside hydrolase family 70 4,6-α-glucanotransferase enzyme (GtfD) to synthesize a reuteran like polymer from maltodextrins and starch. Biochim Biophys Acta. 2016;1860:1224–1236. doi: 10.1016/j.bbagen.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 173.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Machovic M, Janecek S. Starch-binding domains in the post-genome era. Cell Mol Life Sci. 2006;63:2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Carvalho CC, Phan NN, Chen Y, Reilly PJ. Carbohydrate-binding module tribes. Biopolymers. 2015;103:203–214. doi: 10.1002/bip.22584. [DOI] [PubMed] [Google Scholar]
- 176.Christiansen C, Abou Hachem M, Janecek S, Viksø-Nielsen A, Blennow A, Svensson B. The carbohydrate-binding module family 20—diversity, structure, and function. FEBS J. 2009;276:5006–5029. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- 177.Janecek S, Svensson B, MacGregor EA. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb Technol. 2011;49:429–440. doi: 10.1016/j.enzmictec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 178.Majzlova K, Janecek S. Two structurally related starch-binding domain families CBM25 and CBM26. Biologia. 2014;69:1087–1096. doi: 10.2478/s11756-014-0415-3. [DOI] [Google Scholar]
- 179.Koropatkin NM, Smith TJ. SusG: a unique cell-membrane-associated α-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 180.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J Biol Chem. 2012;287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Peng H, Zheng Y, Chen M, Wang Y, Xiao Y, Gao Y. A starch-binding domain identified in α-amylase (AmyP) represents a new family of carbohydrate-binding modules that contribute to enzymatic hydrolysis of soluble starch. FEBS Lett. 2014;588:1161–1167. doi: 10.1016/j.febslet.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 182.Xu J, Ren F, Huang CH, Zheng Y, Zhen J, Sun H, Ko TP, He M, Chen CC, Chan HC, Guo RT, Song H, Ma Y. Functional and structural studies of pullulanase from Anoxybacillus sp. LM18-11. Proteins. 2014;82:1685–1693. doi: 10.1002/prot.24498. [DOI] [PubMed] [Google Scholar]
- 183.Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM, Bayer EA, Flint HJ. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii . MBio. 2015;6:e01058–e01115. doi: 10.1128/mBio.01058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Foley MH, Cockburn DW, Koropatkin NM. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pfister B, Zeeman SC. Formation of starch in plant cells. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Niittyla T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, Gatehouse JA, Villadsen D, Smith SM, Chen J, Zeeman SC, Smith AM. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 187.Minassian BA, Ianzano L, Meloche M, Andermann E, Rouleau GA, Delgado-Escueta AV, Scherer SW. Mutation spectrum and predicted function of laforin in Lafora’s progressive myoclonus epilepsy. Neurology. 2000;55:341–346. doi: 10.1212/WNL.55.3.341. [DOI] [PubMed] [Google Scholar]
- 188.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem Biophys Res Commun. 2004;313:1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 189.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo . Proc Natl Acad Sci USA. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gentry MS, Roma-Mateo C, Sanz P. Laforin, a protein with many faces: glucan phosphatase, adapter protein, et alii. FEBS J. 2013;280:525–537. doi: 10.1111/j.1742-4658.2012.08549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Raththagala M, Brewer MK, Parker MW, Sherwood AR, Wong BK, Hsu S, Bridges TM, Paasch BC, Hellman LM, Husodo S, Meekins DA, Taylor AO, Turner BD, Auger KD, Dukhande VV, Chakravarthy S, Sanz P, Woods VL, Jr, Li S, Vander Kooi CW, Gentry MS. Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease. Mol Cell. 2015;57:261–272. doi: 10.1016/j.molcel.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sankhala RS, Koksal AC, Ho L, Nitschke F, Minassian BA, Cingolani G. Dimeric quaternary structure of human laforin. J Biol Chem. 2015;290:4552–4559. doi: 10.1074/jbc.M114.627406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, Ritte G, Zeeman SC. STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana . Plant Cell. 2009;21:334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. Structural basis for the glucan phosphatase activity of starch excess4. Proc Natl Acad Sci USA. 2010;107:15379–15384. doi: 10.1073/pnas.1009386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Emanuelle S, Brewer MK, Meekins DA, Gentry MS. Unique carbohydrate binding platforms employed by the glucan phosphatases. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hudson ER, Pan DA, James J, Lucocq JM, Hawley SA, Green KA, Baba O, Terashima T, Hardie DG. A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Curr Biol. 2003;13:861–866. doi: 10.1016/S0960-9822(03)00249-5. [DOI] [PubMed] [Google Scholar]
- 199.Polekhina G, Gupta A, Michell BJ, van Denderen B, Murthy S, Feil SC, Jennings IG, Campbell DJ, Witters LA, Parker MW, Kemp BE, Stapleton D. AMPK β subunit targets metabolic stress sensing to glycogen. Curr Biol. 2003;13:867–871. doi: 10.1016/S0960-9822(03)00292-6. [DOI] [PubMed] [Google Scholar]
- 200.Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 201.Emanuelle S, Hossain MI, Moller IE, Pedersen HL, van de Meene AM, Doblin MS, Koay A, Oakhill JS, Scott JW, Willats WG, Kemp BE, Bacic A, Gooley PR, Stapleton DI. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015;82:183–192. doi: 10.1111/tpj.12813. [DOI] [PubMed] [Google Scholar]
- 202.Janecek S. A motif of a microbial starch-binding domain found in human genethonin. Bioinformatics. 2002;18:1534–1537. doi: 10.1093/bioinformatics/18.11.1534. [DOI] [PubMed] [Google Scholar]
- 203.Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, Roach PJ. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Palopoli N, Busi MV, Fornasari MS, Gomez-Casati D, Ugalde R, Parisi G. Starch-synthase III family encodes a tandem of three starch-binding domains. Proteins. 2006;65:27–31. doi: 10.1002/prot.21007. [DOI] [PubMed] [Google Scholar]
- 205.Gomez-Casati DF, Martín M, Busi MV. Polysaccharide-synthesizing glycosyltransferases and carbohydrate binding modules: the case of starch synthase III. Protein Pept Lett. 2013;20:856–863. doi: 10.2174/0929866511320080003. [DOI] [PubMed] [Google Scholar]
- 206.Barchiesi J, Hedin N, Gomez-Casati DF, Ballicora MA, Busi MV. Functional demonstrations of starch binding domains present in Ostreococcus tauri starch synthases isoforms. BMC Res Notes. 2015;8:613. doi: 10.1186/s13104-015-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mikkelsen R, Suszkiewicz K, Blennow A. A novel type carbohydrate-binding module identified in α-glucan, water dikinases is specific for regulated plastidial starch metabolism. Biochemistry. 2006;45:4674–4682. doi: 10.1021/bi051712a. [DOI] [PubMed] [Google Scholar]
- 208.Glaring MA, Baumann MJ, Abou Hachem M, Nakai H, Nakai N, Santelia D, Sigurskjold BW, Zeeman SC, Blennow A, Svensson B. Starch-binding domains in the CBM45 family—low-affinity domains from glucan, water dikinase and α-amylase involved in plastidial starch metabolism. FEBS J. 2011;278:1175–1185. doi: 10.1111/j.1742-4658.2011.08043.x. [DOI] [PubMed] [Google Scholar]
- 209.Hejazi M, Steup M, Fettke J. The plastidial glucan, water dikinase (GWD) catalyses multiple phosphotransfer reactions. FEBS J. 2012;279:1953–1966. doi: 10.1111/j.1742-4658.2012.08576.x. [DOI] [PubMed] [Google Scholar]
- 210.Orzechowski S, Grabowska A, Sitnicka D, Siminska J, Felus M, Dudkiewicz M, Fudali S, Sobczak M. Analysis of the expression, subcellular and tissue localisation of phosphoglucan, water dikinase (PWD/GWD3) in Solanum tuberosum L.: a bioinformatics approach for the comparative analysis of two α-glucan, water dikinases (GWDs) from Solanum tuberosum L. Acta Physiol Plant. 2013;35:483–500. doi: 10.1007/s11738-012-1091-y. [DOI] [Google Scholar]
- 211.Mahlow S, Orzechowski S, Fettke J. Starch phosphorylation: insights and perspectives. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vu VV, Beeson WT, Span EA, Farquhar ER, Marletta MA. A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111:13822–13827. doi: 10.1073/pnas.1408090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lo Leggio L, Simmons TJ, Poulsen JC, Frandsen KE, Hemsworth GR, Stringer MA, von Freiesleben P, Tovborg M, Johansen KS, De Maria L, Harris PV, Soong CL, Dupree P, Tryfona T, Lenfant N, Henrissat B, Davies GJ, Walton PH. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun. 2015;6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vu VV, Marletta MA. Starch-degrading polysaccharide monooxygenases. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cuyvers S, Dornez E, Delcour JA, Courtin CM. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. 2012;32:93–107. doi: 10.3109/07388551.2011.561537. [DOI] [PubMed] [Google Scholar]
- 216.Cockburn D, Wilkens C, Ruzanski C, Andersen S, Willum Nielsen J, Smith AM, Field RA, Willemoës M, Abou Hachem M, Svensson B. Analysis of surface binding sites (SBSs) in carbohydrate active enzymes with focus on glycoside hydrolase families 13 and 77—a mini-review. Biologia. 2014;69:705–712. doi: 10.2478/s11756-014-0373-9. [DOI] [Google Scholar]
- 217.Ragunath C, Manuel SG, Venkataraman V, Sait HB, Kasinathan C, Ramasubbu N. Probing the role of aromatic residues at the secondary saccharide-binding sites of human salivary α-amylase in substrate hydrolysis and bacterial binding. J Mol Biol. 2008;384:1232–1248. doi: 10.1016/j.jmb.2008.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cockburn D, Nielsen MM, Christiansen C, Andersen JM, Rannes JB, Blennow A, Svensson B. Surface binding sites in amylase have distinct roles in recognition of starch structure motifs and degradation. Int J Biol Macromol. 2015;75:338–345. doi: 10.1016/j.ijbiomac.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 219.Suzuki E, Suzuki R. Distribution of glucan-branching enzymes among prokaryotes. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Coutinho PM, Henrissat B. Life with no sugars? J Mol Microbiol Biotechnol. 1999;1:307–308. [PubMed] [Google Scholar]
- 221.UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 223.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 224.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 225.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, Green RK, Flippen-Anderson JL, Westbrook J, Berman HM, Bourne PE. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucl Acids Res. 2005;3:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC
- 227.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 228.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 229.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]