Fig. 5.
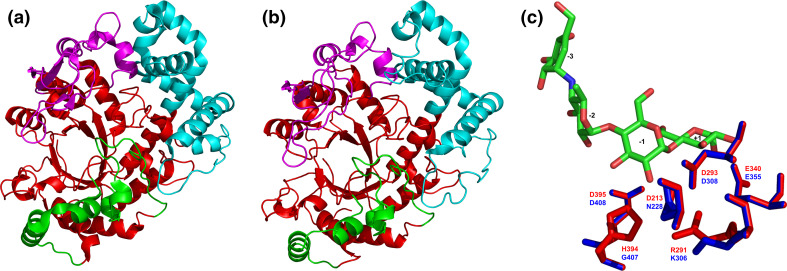
Tertiary structures of the family GH77 amylomaltases from bacteria and their comparison. Sources of the amylomaltases: a Thermus aquaticus [139]; b Borrelia burgdorferi [129]. c Structural overlay focusing on the active-site residues with complexed acarbose in the Thermus amylomaltase (red) compared with the naturally mutated corresponding residues in the Borrelia amylomaltase (blue). The structures were superimposed using the MultiProt web-server [227] (http://bioinfo3d.cs.tau.ac.il/MultiProt/); the overlap being characterized by 488 corresponding Cα-atoms and the RMSD value of 0.18 Å. The Thermus amylomaltase is experimentally solved crystal structure retrieved from the PDB [225] under the PDB code 1ESW, whereas the Borrelia amylomaltase is a tertiary structure model obtained at the homology modeling server SwissModel [228] for its amino acid sequence (UniProt accession number: A6YM39; residues: Asn12-Ala507) based on the T. aquaticus amylomaltase structure (1CWY [44]) as template. The individual domains are colored as follows: catalytic (β/α)8-barrel—red, subdomain B1—cyan, subdomain B2—magenta, subdomain B3—green. The exact positions of displayed active-site residues in the amino acid sequence can be identified in the alignment shown in Fig. 6. Based on the accepted nomenclature [229] the acarbose occupies the subsites from −3 to +1. The structures were visualized with the program PyMol [226]