Fig. 1.
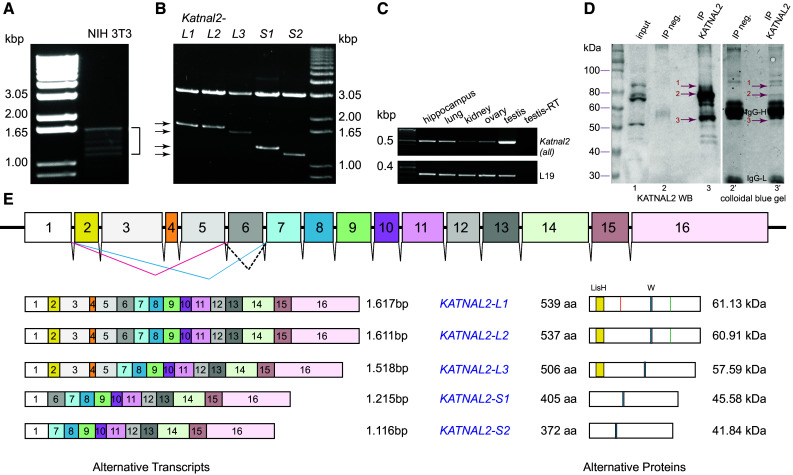
Identification of a family of KATNAL2 isoforms with N-terminal sequence variation. a RT-PCR amplification of cDNA from NIH 3T3 cells, using primers (SET1 in Table S1) at the extremities of the predicted ORF of mouse Katnal2 XM_006526437. Several distinct bands (1.2–1.6 kbp) were obtained (bracket). b The mixed PCR product from (a) was subcloned in vector pGEM T easy, and after screening of 100 clones, five distinct types of cDNA, shown here as products of SacII/SacI restriction digests (arrows), were identified by DNA sequencing and assigned names Katnal2-L1, -L2, -L3, -S1 and -S2. c Semi-quantitative RT-PCR analysis of Katnal2 expression in selected mouse tissues, using primers (SET5 in Table S1) that resulted in the amplification of a diagnostic product of 508 bp, common to all Katnal2 transcripts. d Analysis of IP experiment from mouse testis, using the affinity purified anti-KATNAL2 antibody. IP samples were analyzed in parallel by WB (20 %, left) and on a colloidal-blue-stained gel (80 %, right) to identify unique bands in the KATNAL2-IP that were immunoreactive. Three such bands were identified (red arrows bands 1–3) and cut out for MS analysis. Lanes 1, 1′ input; 2, 2′ negative control IP (goat IgG); 3, 3′ KATNAL2 IP. Results from MS analysis in Table S3, confirm the presence of KATNAL2 proteins in these immunoreactive bands. e Sketch of the Katnal2 gene on mouse chromosome 18. Exons (1–16) are colour coded and to scale (introns are not to scale). The five distinct cDNAs in b correspond to transcripts designated Katnal2-L1, L2, L3, S1 and S2 with differential exon combinations, as shown on the left of the panel. Corresponding encoded proteins are shown on the right-hand side; isoforms L1 and L2 differ only in two additional amino acids (VK) in L1; their position is marked by a red line in L1. Protein motifs are shown: LisH domain as a yellow box, Walker motif as a blue box and SNPs in L1 and L2 (H instead of R) are indicated as a green line. Full protein sequences in Fig. S1. cDNA sequences were deposited to the European Nucleotide Archive (www.ebi.ac.uk/ena/data/view/LN831862-LN831866)
