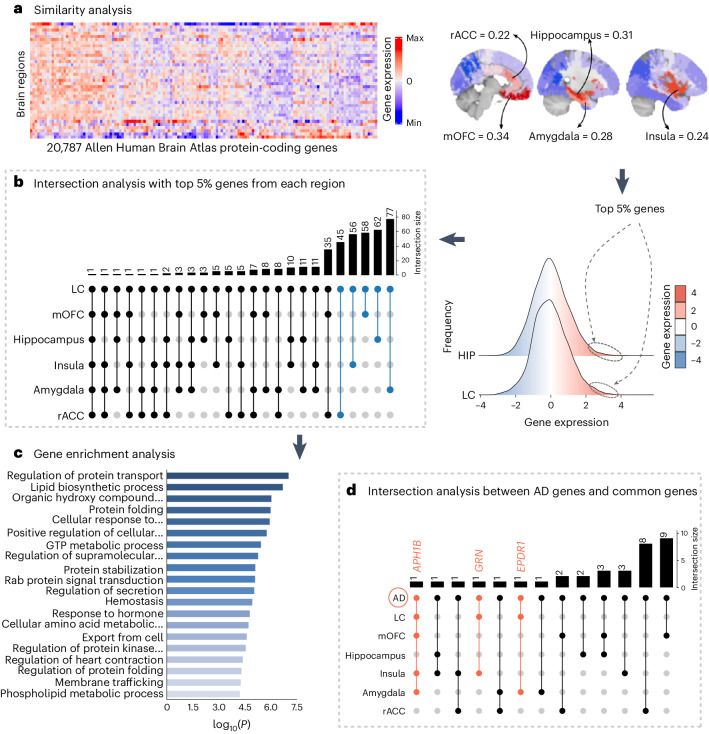
Fig. 4. Neurogenetic approach exploring common genetic background across the brain.
a, A whole-brain region-wise phenotypic-transcriptomic similarity analysis using the AHBA was conducted correlating LC to 68 regions from the Desikan–Killiany neocortical parcellation, the hippocampus and the amygdala. The sagittal brain slices (FSLeyes; FSL, FMRIB) show LC’s gene expression profile similarity to that of the hippocampus, amygdala, insula, mOFC and rACC (warmer colors indicate higher similarity). The genes with the highest genetic expression (top 5%) within these six regions were selected for subsequent analysis (the distribution corresponding to the genetic expression of protein-coding genes colocated at the LC and the hippocampus are shown). b, An intersection analysis was used to define common protein-coding genes between LC and each of the other regions, aggregating genes involving LC plus one region (n = 298 protein-coding genes). c, GO enrichment analysis revealed that among the biological functions related to these genes, which are highly expressed in early AD-affected regions, regulation of protein transport was found to be the main term (thresholds for terms, P < 0.01, count > 3 and enrichment factor > 1.5; P values, log10(P) are calculated based on the cumulative hypergeometric). d, An intersection analysis was used to find, within the common gene expression profiles (n = 298 protein-coding genes), genes related to AD (n = 75 genes). The APH1B, GRN and EPDR1 genes were found within the AD-related genes and our neurogenetic approach results. GTP, guanosine triphosphate; HIPP, hippocampus.