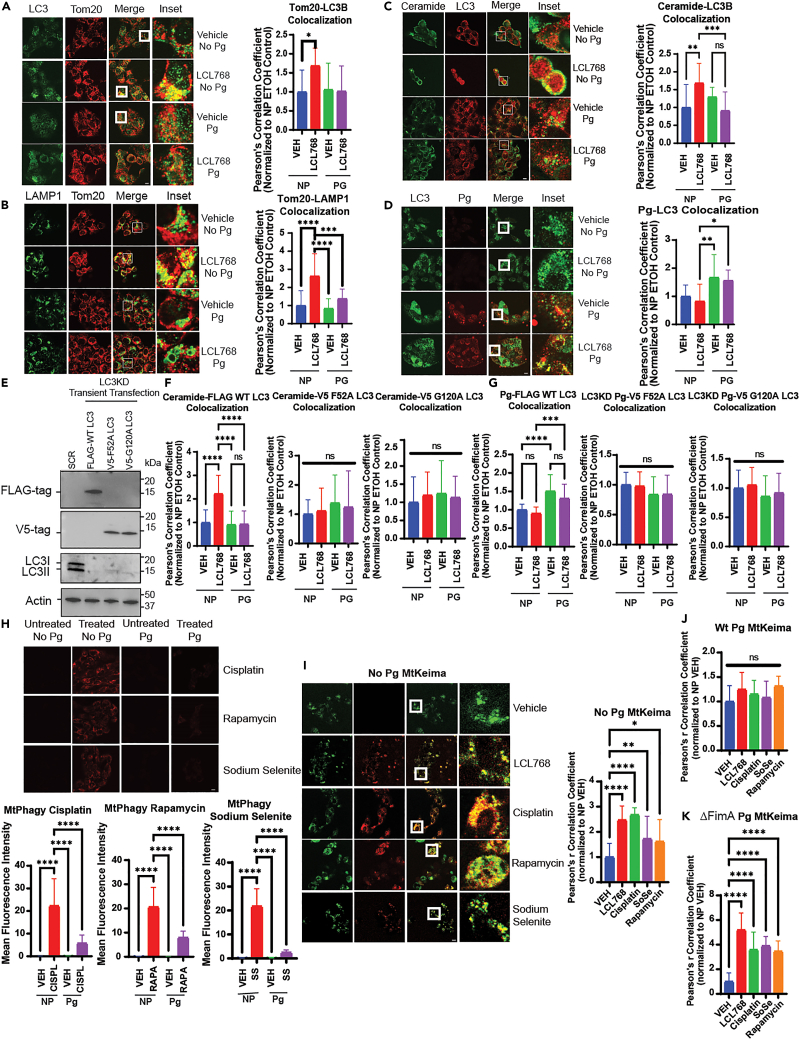
Figure 1.
P gingivalis inhibits mitophagy through disruption of ceramide, LC3, and Annexin A2 (ANXA2) binding
(A) Confocal images of UMSCC1A cells infected/uninfected with P. gingivalis (100 MOI, 6 h) and treated with vehicle or LCL768 (30 μM, 2 h) were stained for LC3 (green) and Tom20 (red).
(B) Confocal images of UMSCC1A cells infected/uninfected with P. gingivalis and treated with vehicle or LCL768, labeled for LAMP1 (green) and Tom20 (red).
(C and D) Similarly, confocal images of ceramide (green) and LC3 (red), and LC3 (green) and P. gingivalis (red) are shown. Yellow shows colocalization. Images represent three independent experiments. Scale bars are 100 μm (throughout the manuscript unless specifically noted). Quantification of colocalization was estimated via Pearson’s correlation coefficient, completed with ImageJ Fiji software, and normalized to uninfected, untreated control. Values indicate mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(E) Western blotting was used to detect the protein abundance of stably knocked down LC3 and transiently transfected tagged mutant or wildtype LC3 in UMSCC1A cells. Actin was used as the loading control.
(F) UMSCC1A cells stably expressing shRNA against LC3 and transiently transfected with FLAG-wildtype LC3, V5-F52A LC3, or V5-G120A LC3 pcDNA3.1+ plasmids were labeled with ceramide antibody and tag antibody (FLAG or V5) for confocal microscopy. Quantification of colocalization was estimated using Pearson’s correlation coefficient and normalized to uninfected, untreated control using ImageJ Fiji software. Images represent three independent experiments. Data are means ± SD (n = 3, ns, not significant, ∗∗∗∗p < 0.0001).
(G) UMSCC1A cells stably expressing shRNA against LC3 were transiently transfected with FLAG-wildtype LC3, V5-F52A LC3, or V5-G120A LC3 plasmids before infection with P. gingivalis (6 h, 100 MOI) and treatment with 30 μM LCL768 or vehicle for 3 h. Cells were dual labeled with P. gingivalis antibody and the appropriate tag antibody (FLAG or V5) for confocal microscopy. Quantification of colocalization, shown via Pearson’s correlation coefficient, are means ± SD (n = 3, ns, not significant, ∗∗∗∗p < 0.0001).
(H) Live UMSCC1A cells were stained using Mtphagy dye (red) in which fluorescence intensity increases with mitophagy induction, following treatment with 5 μM cisplatin, 8 μM rapamycin, or 8 μM sodium selenite for 3 h. Images represent three independent experiments. The bottom panels show the quantification of Mtphagy fluorescence intensity using ImageJ Fiji software. Data are means ± SD (n = 3, ∗∗∗∗p < 0.0001).
(I) UMSCC1A cells were transiently transfected with pHAGE-MtKeima plasmid, in which auto-fluorescent Keima protein conjugated to mitochondrial-targeting sequence (COX8) fluoresces green at neutral pH (>pH 6) and red at acidic pH (<pH 5). Live cells were then treated (2 h) with vehicle control, LCL768, or alternative mitophagy-inducing drugs following infection/no infection with P. gingivalis (6 h). Images represent three independent experiments.
(J and K) Quantification of colocalization in cells infected with wildtype or FimA deletion mutant P. gingivalis (ΔFimA) are shown via Pearson’s correlation coefficient as means ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).