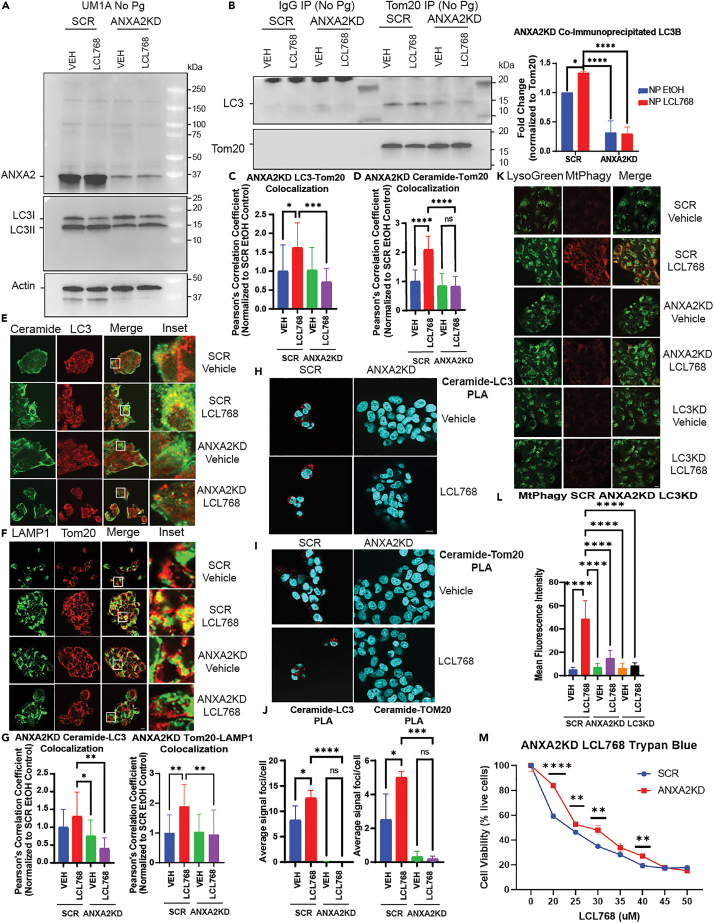
Figure 3.
ANXA2 knockdown inhibits mitophagy
(A and B) Immunoprecipitation with Tom20 or species-matched IgG antibody in uninfected UMSCC1A cells stably expressing shRNA against ANXA2 or scrambled control. The left panel shows input (pre-immunoprecipitation) ANXA2 and LC3 via western blotting, normalized to Actin control bands. The right panel shows co-immunoprecipitated LC3 in cells treated with LCL768 or vehicle control. Images represent at least three independent experiments. Quantification of co-immunoprecipitated LC3 western blot bands was normalized to scrambled shRNA expressing, untreated control. Data are means ± SD (n = 3, ∗p < 0.05, ∗∗∗∗p < 0.0001).
(C and D) Quantification of confocal images of uninfected UMSCC1A cells stably expressing shRNA against ANXA2 or scrambled control and treated with LCL768 or vehicle control. Colocalization between LC3 and Tom20 (C), or ceramide and Tom20 (D), was estimated via Pearson’s correlation coefficient and normalized to uninfected, untreated control. Values indicate mean ± SD of n = 3 independent experiments. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(E and F) Confocal images of uninfected UMSCC1A cells stably expressing shRNA against ANXA2 or scrambled control, treated with LCL768 or vehicle control and dual labeled with ceramide (green) and LC3 (red), or LAMP1 (green) and Tom20 (red) antibodies.
(G) Quantification of confocal images in (E) and (F), shown via Pearson’s correlation coefficient. Values indicate mean ± SD of n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01.
(H and I) Proximity ligation assay was performed on uninfected UMSCC1A cells stably expressing shRNA against ANXA2 or scrambled control for ceramide and LC3, or ceramide and Tom20. Confocal images show DAPI nuclei staining (blue) and PLA signal (red) indicating interaction of targets within close proximity (<40 nm).
(J) Quantification of the average number of PLA signal foci per cell in (H) and (I) was completed with ImageJ Fiji. Values indicate mean ± SD of n = 3 independent experiments. Ns, not significant, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(K) Live, uninfected UMSCC1A cells stably expressing shRNA against ANXA2, LC3, or scrambled control were stained with Mtphagy (red) and LysoGreen dyes following treatment with LCL768 or vehicle control. Images represent three independent experiments. Yellow indicates colocalization.
(L) Quantification of Mtphagy fluorescence intensity in (K) was done using ImageJ Fiji software. Data are means ± SD (n = 3, ∗∗∗∗p < 0.0001).
(M) Live, uninfected UMSCC1A cells stably expressing shRNA against ANXA2, or scrambled control, were treated with increasing concentrations of LCL768 for 2 h and stained with trypan blue cell exclusion dye. Cell viability (percent live, unstained cells) is shown as means ± SD (n = 3, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).