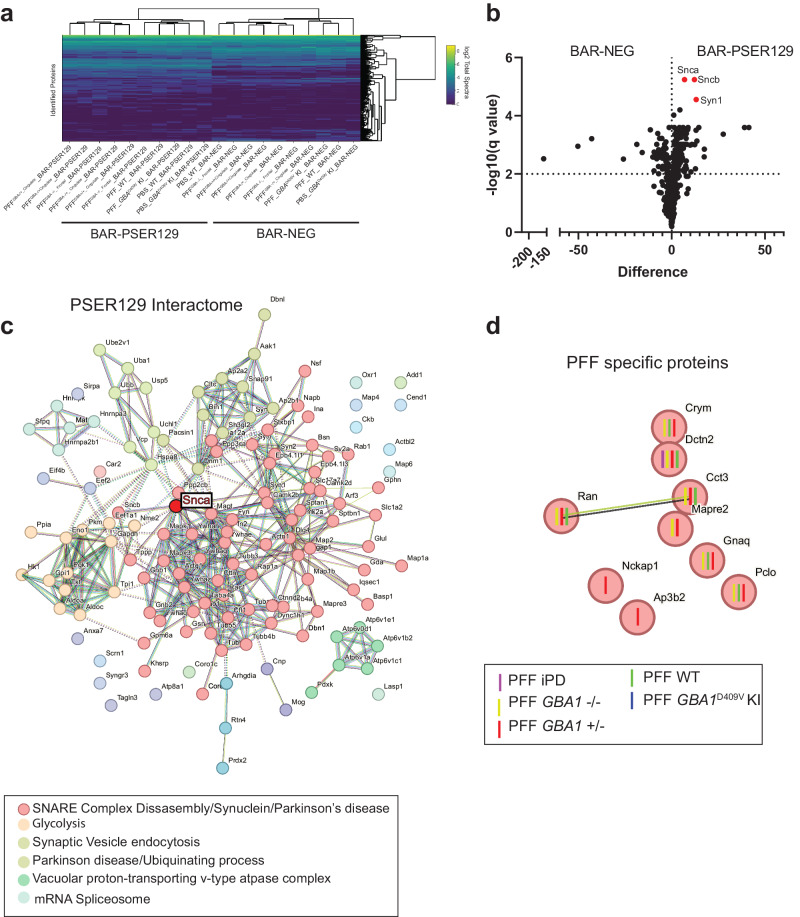
Fig. 5. Identified PSER129 interactome in the brain of mice under different seeding conditions.
BAR-PSER129 was performed on pooled (4-5 mice) brain samples from each experimental group. LC-MS/MS identified a total of 464 proteins. a Unbiased hierarchical clustering of identified proteins. Two major clusters were observed according to capture conditions and annotated “BAR-PSER129” and “BAR-NEG.” b Volcano plot of all proteins identified. BAR-PSER129 and BAR-NEG were compared. Top hits, Alpha- (Snca), beta-synuclein (Sncb), and synapsin I (Syn1) were denoted in red and labeled. c STRING interaction map of 144 proteins significantly enriched over background (BAR-NEG). MCL clustering (inflation parameter 1.4) grouped proteins into 6 functional clusters. Most populated clusters are annotated with consensus terms for significantly enriched pathways. The position of αsyn (Snca) is highlighted. d PSER129-interacting proteins identified exclusively in brains of PFF seeded mice. Each protein is annotated with the seeding conditions for which the protein was identified. “PFF iPD”, “PFF GBA1 -/-”, “PFF GBA1 +/-” are PFF strains derived from PD cases without GBA mutations, homozygous, and heterozygous mutations, respectively. “PFF WT” and “PFF GBA1D409V KI” were WT and GBA1 D409V KI mice, respectively, seeded with de novo PFFs.