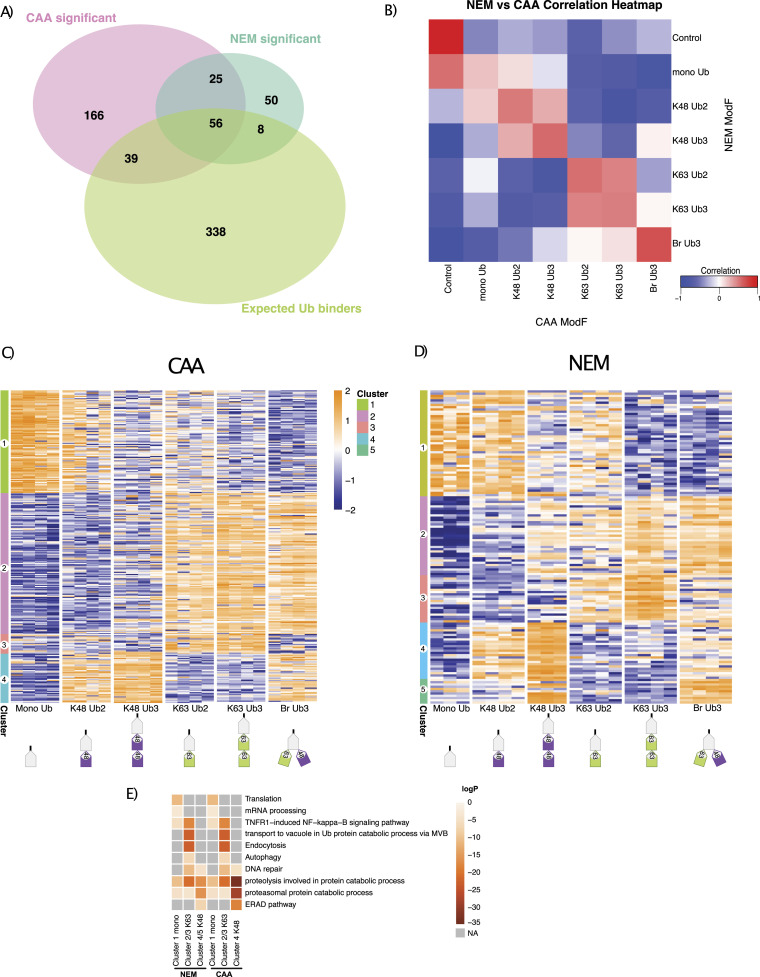
Figure 2. Comparison of interactor enrichment patterns between chloroacetamide (CAA) and N-ethylmaleimide (NEM) datasets.
(A) Overlap of significant differently enriched interactors across Ub pulldowns from the CAA- and NEM-treated lysate with expected Ub binders (expected UbBPs). Data were prefiltered by Ub enrichment (significant enrichment on Ub over bead-only control in at least one two-sample moderated t test comparison, log(FC) > 0, Adj.P < 0.05). Significant differently enriched interactors were identified from prefiltered Ub-enriched interactors by a moderated F test (Adj.P < 0.05). The expected Ub binder list was compiled from the Gene Ontology term Ub-binding 0043130 and UBD-containing proteins from the iUUCD (38) (http://iuucd.biocuckoo.org/) and literature research. (B) Correlation of bait interactomes between CAA and NEM datasets. The Spearman correlation was calculated by comparing the moderated F values of common significant differently enriched proteins in each pulldown between datasets. A moderated F test before prefiltering (Adj.P < 0.05). (C, D) Clustering of significant differently enriched proteins from (C) CAA and (D) NEM datasets. (A) Significance determination as in (A). Hierarchical clustering by the Euclidean distance. Heatmap of iBAQ values, scaled by row using z scoring. Large-scale version of heatmaps including protein row names in Figs S5 and S6. (C, D, E) Gene Ontology (GO) enrichment heatmap of clusters from (C, D). GO enrichment was calculated with Metascape (39) (http://metascape.org), with a minimum overlap of 2, P-value cut-off < 0.01, and minimum enrichment of 1.5. Grey is not enriched. Some GO terms are abbreviated: TNFR1−induced NF−kappa−B is TNFR1−induced NF−kappa−B signalling pathway, transport to the vacuole in Ub protein catabolic process via MVB is protein transport to the vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway, and ERAD is endoplasmic reticulum–associated protein degradation.